5JFY
 
 | |
1JDI
 
 | | CRYSTAL STRUCTURE OF L-RIBULOSE-5-PHOSPHATE 4-EPIMERASE | | Descriptor: | L-RIBULOSE 5 PHOSPHATE 4-EPIMERASE, ZINC ION | | Authors: | Luo, Y, Samuel, J, Mosimann, S.C, Lee, J.E, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 2001-06-13 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of L-ribulose-5-phosphate 4-epimerase: an aldolase-like platform for epimerization.
Biochemistry, 40, 2001
|
|
1K0W
 
 | | CRYSTAL STRUCTURE OF L-RIBULOSE-5-PHOSPHATE 4-EPIMERASE | | Descriptor: | L-RIBULOSE 5 PHOSPHATE 4-EPIMERASE, ZINC ION | | Authors: | Luo, Y, Samuel, J, Mosimann, S.C, Lee, J.E, Strynadka, N.C.J. | | Deposit date: | 2001-09-21 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of L-ribulose-5-phosphate 4-epimerase: an aldolase-like platform for epimerization
Biochemistry, 40, 2001
|
|
3MMS
 
 | | Crystal structure of Streptococcus pneumoniae MTA/SAH nucleosidase in complex with 8-aminoadenine | | Descriptor: | 5'-methylthioadenosine / S-adenosylhomocysteine nucleosidase, 9H-purine-6,8-diamine, GLYCEROL | | Authors: | Siu, K.K.W, Lee, J.E, Horvatin-Mrakovcic, C, Howell, P.L. | | Deposit date: | 2010-04-20 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae MTA/SAH nucleosidase in complex with 8-aminoadenine
TO BE PUBLISHED
|
|
7N05
 
 | |
7N07
 
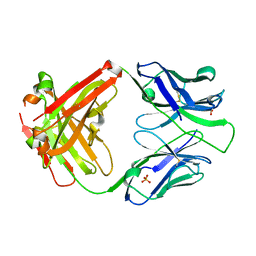 | | Crystal structure of the apo 3D6 antibody fragment | | Descriptor: | Fab 3D6 heavy chain, Fab 3D6 light chain, SULFATE ION | | Authors: | Cook, J.D, Lee, J.E. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational plasticity of the HIV-1 gp41 immunodominant region is recognized by multiple non-neutralizing antibodies.
Commun Biol, 5, 2022
|
|
7N04
 
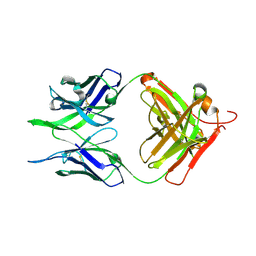 | | Crystal structure of the apo F240 antibody fragment | | Descriptor: | Fab F240 heavy chain, Fab F240 light chain, alpha-D-glucopyranose | | Authors: | Cook, J.D, Lee, J.E. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.70000517 Å) | | Cite: | Conformational plasticity of the HIV-1 gp41 immunodominant region is recognized by multiple non-neutralizing antibodies.
Commun Biol, 5, 2022
|
|
7N08
 
 | |
3TIS
 
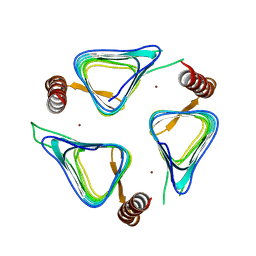 | | Crystal structures of yrdA from Escherichia coli, a homologous protein of gamma-class carbonic anhydrases, show possible allosteric conformations | | Descriptor: | Protein YrdA, ZINC ION | | Authors: | Park, H.M, Chio, J.W, Lee, J.E, Jung, J.H, Kim, B.Y, Kim, J.S. | | Deposit date: | 2011-08-21 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the gamma-class carbonic anhydrase homologue YrdA suggest a possible allosteric switch
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TIO
 
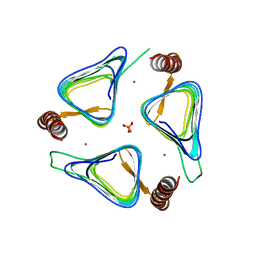 | | Crystal structures of yrdA from Escherichia coli, a homologous protein of gamma-class carbonic anhydrase, show possible allosteric conformations | | Descriptor: | PHOSPHATE ION, Protein YrdA, ZINC ION | | Authors: | Park, H.M, Choi, J.W, Lee, J.E, Jung, C.H, Kim, B.Y, Kim, J.S. | | Deposit date: | 2011-08-21 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structures of the gamma-class carbonic anhydrase homologue YrdA suggest a possible allosteric switch
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5UQY
 
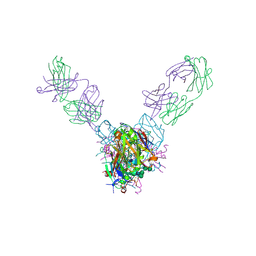 | | Crystal structure of Marburg virus GP in complex with the human survivor antibody MR78 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP1, ... | | Authors: | Hashiguchi, T, Fusco, M.L, Hastie, K.M, Bomholdt, Z.A, Lee, J.E, Flyak, A.I, Matsuoka, R, Kohda, D, Yanagi, Y, Hammel, M, Crowe, J.E, Saphire, E.O. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for Marburg virus neutralization by a cross-reactive human antibody.
Cell, 160, 2015
|
|
4J4J
 
 | |
4JF3
 
 | |
4JGS
 
 | |
4JPR
 
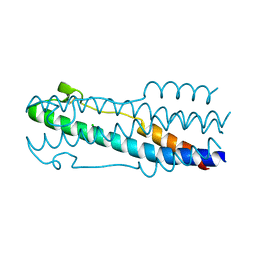 | | Structure of the ASLV fusion subunit core | | Descriptor: | ASLV fusion TM, CHLORIDE ION | | Authors: | Aydin, H, Smrke, B.M, Lee, J.E. | | Deposit date: | 2013-03-19 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural characterization of a fusion glycoprotein from a retrovirus that undergoes a hybrid 2-step entry mechanism.
Faseb J., 27, 2013
|
|
5F4E
 
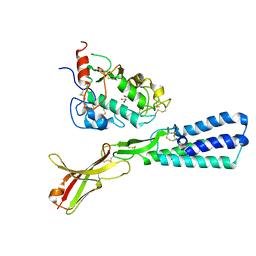 | | Crystal structure of the human sperm Izumo1 and egg Juno complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Aydin, H, Sultana, A, Lee, J.E. | | Deposit date: | 2015-12-03 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex.
Nature, 534, 2016
|
|
5F4V
 
 | |
5F4Q
 
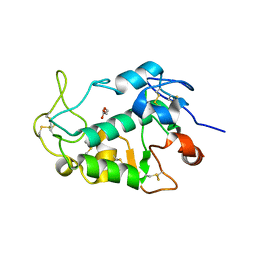 | | Crystal structure of the human egg surface protein Juno | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Aydin, H, Sultana, A, Lee, J.E. | | Deposit date: | 2015-12-03 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex.
Nature, 534, 2016
|
|
5F4T
 
 | |
5T9Y
 
 | | Crystal structure of the infectious salmon anemia virus (ISAV) hemagglutinin-esterase protein | | Descriptor: | FORMIC ACID, GLYCEROL, HE protein, ... | | Authors: | Cook, J.D, Sultana, A, Lee, J.E. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-22 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the infectious salmon anemia virus receptor complex illustrates a unique binding strategy for attachment.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T96
 
 | | Crystal structure of the infectious salmon anemia virus (ISAV) HE viral receptor complex | | Descriptor: | 4-O-acetyl-5-acetamido-3,5-dideoxy-L-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Cook, J.D, Sultana, A, Lee, J.E. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the infectious salmon anemia virus receptor complex illustrates a unique binding strategy for attachment.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6KNR
 
 | | Crystal structure of Estrogen-related receptor gamma ligand-binding domain with DN200699 | | Descriptor: | (E)-4-(1-(4-(1-cyclopropylpiperidin-4-yl)phenyl)-5-hydroxy-2-phenylpent-1-en-1-yl)phenol, Estrogen-related receptor gamma | | Authors: | Yoon, H, Kim, J, Chin, J, Song, J, Cho, S.J. | | Deposit date: | 2019-08-07 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | An orally available inverse agonist of estrogen-related receptor gamma showed expanded efficacy for the radioiodine therapy of poorly differentiated thyroid cancer.
Eur.J.Med.Chem., 205, 2020
|
|
6JLA
 
 | | Crystal structure of a mouse ependymin related protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Mammalian ependymin-related protein 1 | | Authors: | Park, S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-04 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of three ependymin-related proteins suggest their function as a hydrophobic molecule binder.
Iucrj, 6, 2019
|
|
6JLD
 
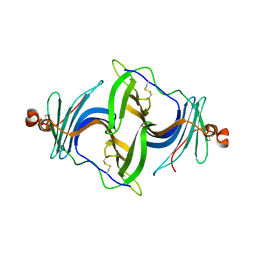 | | Crystal structure of a human ependymin related protein | | Descriptor: | Mammalian ependymin-related protein 1 | | Authors: | Park, S.Y. | | Deposit date: | 2019-03-05 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of three ependymin-related proteins suggest their function as a hydrophobic molecule binder.
Iucrj, 6, 2019
|
|
6JL9
 
 | | Crystal structure of a frog ependymin related protein | | Descriptor: | CALCIUM ION, Ependymin-related 1 | | Authors: | Park, S.Y. | | Deposit date: | 2019-03-04 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of three ependymin-related proteins suggest their function as a hydrophobic molecule binder.
Iucrj, 6, 2019
|
|
