1MNL
 
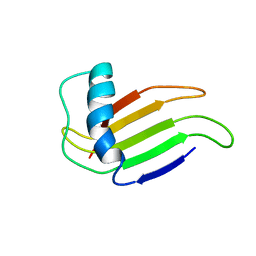 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF A SWEET PROTEIN SINGLE-CHAIN MONELLIN (SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND DYNAMICAL SIMULATED ANNEALING CALCULATIONS, 21 STRUCTURES | | Descriptor: | MONELLIN | | Authors: | Lee, S.-Y, Lee, J.-H, Chang, H.-J, Jo, J.-M, Jung, J.-W, Lee, W. | | Deposit date: | 1998-08-06 | | Release date: | 1999-06-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a sweet protein single-chain monellin determined by nuclear magnetic resonance and dynamical simulated annealing calculations.
Biochemistry, 38, 1999
|
|
4GQZ
 
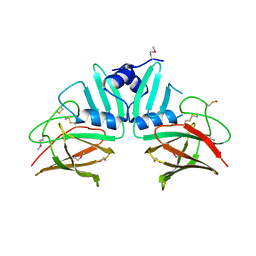 | | Crystal Structure of S.CueP | | Descriptor: | CHLORIDE ION, Putative periplasmic or exported protein | | Authors: | Ha, N.C, Yoon, B.Y. | | Deposit date: | 2012-08-24 | | Release date: | 2013-08-14 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structure of the periplasmic copper-binding protein CueP from Salmonella enterica serovar Typhimurium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5Y3D
 
 | |
4ES0
 
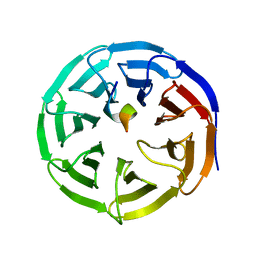 | | X-ray structure of WDR5-SETd1b Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase SETD1B, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.817 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERZ
 
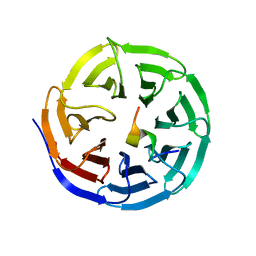 | | X-ray structure of WDR5-MLL4 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL4, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
7CQ0
 
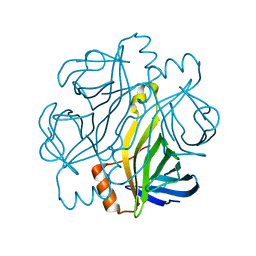 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | Descriptor: | Mature Streptoavidin-C1 | | Authors: | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | Deposit date: | 2020-08-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
7CPZ
 
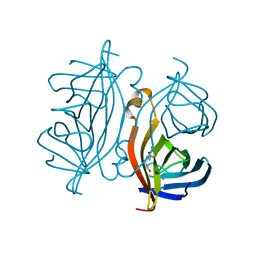 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | Descriptor: | BIOTIN, Mature Streptoavidin-C1 | | Authors: | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | Deposit date: | 2020-08-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
3LDI
 
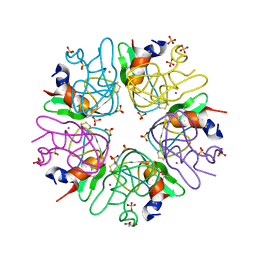 | | Crystal structure of aprotinin in complex with sucrose octasulfate: unusual interactions and implication for heparin binding | | Descriptor: | GLYCEROL, MERCURY (II) ION, Pancreatic trypsin inhibitor, ... | | Authors: | Yang, I.S, Kim, T.G, Park, B.S, Kim, K.H. | | Deposit date: | 2010-01-13 | | Release date: | 2010-06-23 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of aprotinin and its complex with sucrose octasulfate reveal multiple modes of interactions with implications for heparin binding
Biochem.Biophys.Res.Commun., 397, 2010
|
|
1JO7
 
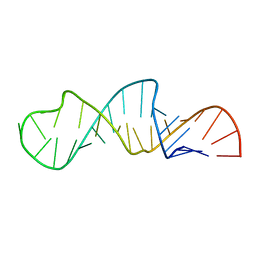 | | Solution Structure of Influenza A Virus Promoter | | Descriptor: | Influenza A virus promoter RNA | | Authors: | Bae, S.-H, Cheong, H.-K, Lee, J.-H, Cheong, C, Kainosho, M, Choi, B.-S. | | Deposit date: | 2001-07-27 | | Release date: | 2001-09-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural features of an influenza virus promoter and their implications for viral RNA synthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
