1EHE
 
 | |
6XOS
 
 | |
6XOU
 
 | |
6XOV
 
 | |
6XOT
 
 | |
6V5D
 
 | | EROS3 RDC and NOE Derived Ubiquitin Ensemble | | Descriptor: | Ubiquitin | | Authors: | Lange, O.F, Lakomek, N.A, Smith, C.A, Griesinger, C, de Groot, B.L. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Enhancing NMR derived ensembles with kinetics on multiple timescales.
J.Biomol.Nmr, 74, 2020
|
|
3U4E
 
 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain CAP45 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG9 Heavy Chain, PG9 Light Chain, ... | | Authors: | Gorman, J, McLellan, J, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3U2S
 
 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain ZM109 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | McLellan, J.S, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3U46
 
 | | CH04H/CH02L P212121 | | Descriptor: | CH02 Light chain Fab, CH04 Heavy chain Fab | | Authors: | Louder, R, Pancera, M, McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
8HRQ
 
 | |
8HRT
 
 | |
8HRP
 
 | |
8HRR
 
 | |
8HRO
 
 | |
8HRS
 
 | |
4TN5
 
 | |
4BBW
 
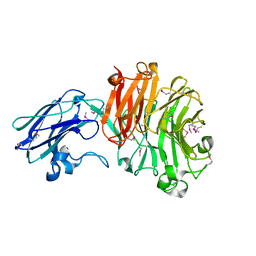 | | The crystal structure of Sialidase VPI 5482 (BTSA) from Bacteroides thetaiotaomicron | | Descriptor: | SIALIDASE (NEURAMINIDASE) | | Authors: | Park, K.-H, Song, H.-N, Jung, T.-Y, Lee, M.-H, Woo, E.-J. | | Deposit date: | 2012-09-28 | | Release date: | 2013-08-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of the Broad Substrate Specificity of Bacteroides Thetaiotaomicron Commensal Sialidase.
Biochim.Biophys.Acta, 1834, 2013
|
|
4B04
 
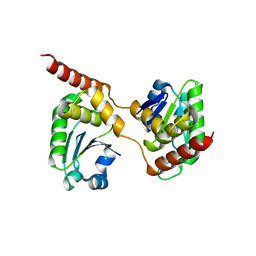 | | Crystal structure of the Catalytic Domain of Human DUSP26 (C152S) | | Descriptor: | DUAL SPECIFICITY PROTEIN PHOSPHATASE 26 | | Authors: | Won, E.-Y, Lee, D.Y, Park, S.G, Yokoyama, S, Kim, S.J, Chi, S.-W. | | Deposit date: | 2012-06-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | High-Resolution Crystal Structure of the Catalytic Domain of Human Dual-Specificity Phosphatase 26
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1BO0
 
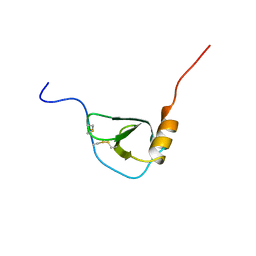 | | MONOCYTE CHEMOATTRACTANT PROTEIN-3, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN (MONOCYTE CHEMOATTRACTANT PROTEIN-3) | | Authors: | Kwon, D, Lee, D, Sykes, B.D, Kim, K.-S. | | Deposit date: | 1998-08-10 | | Release date: | 1999-10-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of a monomeric chemokine: monocyte chemoattractant protein-3.
FEBS Lett., 395, 1996
|
|
4RMK
 
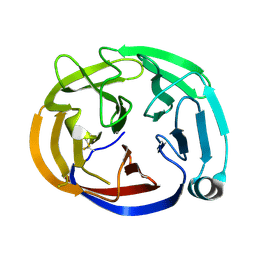 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in P65 crystal form | | Descriptor: | CALCIUM ION, Latrophilin-3 | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-08-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
4RML
 
 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in C2221 crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Latrophilin-3, MAGNESIUM ION | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-08-19 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
7VQI
 
 | |
7VQH
 
 | |
5TZ3
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 2A IN COMPLEX with [1,2,4]triazolo[1,5-a]pyrimidin-7-yl}-N-(naphthalene-2-yl)piperidine-3-carboxamide | | Descriptor: | (3~{R})-1-(5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)-~{N}-naphthalen-2-yl-piperidine-3-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Xu, R, Aertgeerts, K. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Design and Synthesis of Novel and Selective Phosphodiesterase 2 (PDE2a) Inhibitors for the Treatment of Memory Disorders.
J. Med. Chem., 60, 2017
|
|
5U00
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 2A IN COMPLEX WITH 3,3-difluoro-1-[(4-fluoro-3-iodophenyl)carbonyl]-5-{5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-yl}piperidine | | Descriptor: | MAGNESIUM ION, ZINC ION, [(5S)-3,3-difluoro-5-(5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)piperidin-1-yl](4-fluoro-3-iodophenyl)methanone, ... | | Authors: | Xu, R, Aertgeerts, K. | | Deposit date: | 2016-11-22 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Design and Synthesis of Novel and Selective Phosphodiesterase 2 (PDE2a) Inhibitors for the Treatment of Memory Disorders.
J. Med. Chem., 60, 2017
|
|
