8EE8
 
 | |
8EF2
 
 | |
8EF3
 
 | |
5UIS
 
 | | Crystal structure of IRAK4 in complex with compound 12 | | Descriptor: | 4-{[(3R)-piperidin-3-yl]oxy}-6-[(propan-2-yl)oxy]quinoline-7-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIQ
 
 | | Crystal structure of IRAK4 in complex with compound 9 | | Descriptor: | 2-[(propan-2-yl)oxy]benzamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIR
 
 | | Crystal structure of IRAK4 in complex with compound 11 | | Descriptor: | 5-(4-cyanophenyl)-3-[(propan-2-yl)oxy]naphthalene-2-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIT
 
 | | Crystal structure of IRAK4 in complex with compound 14 | | Descriptor: | 1-{[(2S)-5-oxopyrrolidin-2-yl]methoxy}-7-[(propan-2-yl)oxy]isoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIU
 
 | | Crystal structure of IRAK4 in complex with compound 30 | | Descriptor: | 1-{[(2S,3S,4S)-3-ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
6ICI
 
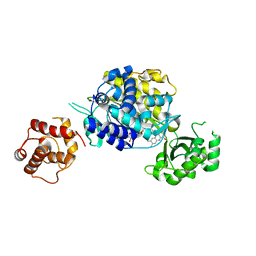 | | Crystal structure of human MICAL3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, [F-actin]-monooxygenase MICAL3 | | Authors: | Hwang, K.Y, Kim, J.S. | | Deposit date: | 2018-09-06 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic insights into flavin-containing monooxygenase and calponin-homology domains in human MICAL3.
Iucrj, 7, 2020
|
|
6ZGC
 
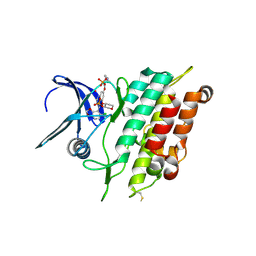 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Saracatinib (AZD0530) | | Descriptor: | Activin receptor type I, N-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-7-[2-(4-METHYLPIPERAZIN-1-YL)ETHOXY]-5-(TETRAHYDRO-2H-PYRAN-4-YLOXY)QUINAZOLIN-4-AMINE, PHOSPHATE ION, ... | | Authors: | Williams, E.P, Galan Bartual, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Saracatinib is an efficacious clinical candidate for fibrodysplasia ossificans progressiva.
JCI Insight, 6, 2021
|
|
4TSM
 
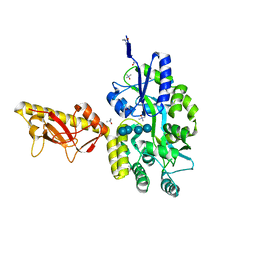 | | MBP-fusion protein of PilA1 from C. difficile R20291 residues 26-166 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein, pilin chimera, ... | | Authors: | Piepenbrink, K.H, Sundberg, E.J. | | Deposit date: | 2014-06-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural and Evolutionary Analyses Show Unique Stabilization Strategies in the Type IV Pili of Clostridium difficile.
Structure, 23, 2015
|
|
5C0N
 
 | |
5D8J
 
 | |
2DF6
 
 | |
3DHX
 
 | | Crystal structure of isolated C2 domain of the methionine uptake transporter | | Descriptor: | IODIDE ION, Methionine import ATP-binding protein metN | | Authors: | Johnson, E, Kaiser, J.T, Lee, A.T, Rees, D.C. | | Deposit date: | 2008-06-18 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation.
Science, 321, 2008
|
|
3DHW
 
 | | Crystal structure of methionine importer MetNI | | Descriptor: | D-methionine transport system permease protein metI, Methionine import ATP-binding protein metN | | Authors: | Rees, D.C, Kaiser, J.T, Kadaba, N.S, Johnson, E, Lee, A.T. | | Deposit date: | 2008-06-18 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation.
Science, 321, 2008
|
|
3KXZ
 
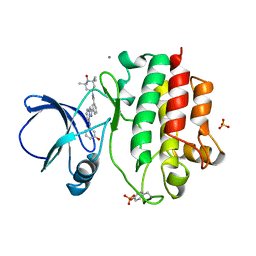 | | The complex crystal structure of LCK with a probe molecule w259 | | Descriptor: | 3-[7-[(3-hydroxyphenyl)amino]pyrazolo[1,5-a]pyrimidin-2-yl]-N-(1-hydroxy-2,2,6,6-tetramethyl-piperidin-4-yl)benzamide, CALCIUM ION, Proto-oncogene tyrosine-protein kinase LCK, ... | | Authors: | Xu, Z.B, Moy, F.J, Kelleher, K, Mosyak, L, Protein Structure Factory (PSF) | | Deposit date: | 2009-12-04 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Novel synthesis and structural characterization of a high-affinity paramagnetic kinase probe for the identification of non-ATP site binders by nuclear magnetic resonance.
J.Med.Chem., 53, 2010
|
|
5KJF
 
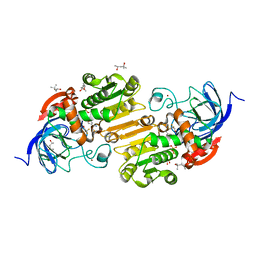 | | V222I horse liver alcohol dehydrogenase complexed with NAD+ and trifluoroethanol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alcohol dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), ... | | Authors: | Plapp, B.V. | | Deposit date: | 2016-06-18 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Contribution of buried distal amino acid residues in horse liver alcohol dehydrogenase to structure and catalysis.
Protein Sci., 27, 2018
|
|
5KJ6
 
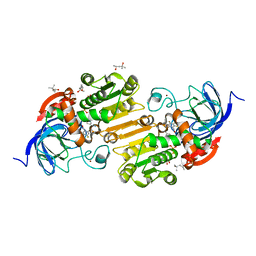 | | V197I Horse liver alcohol dehydrogenase complexed with NAD+ and pentafluorobenzyl alcohol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Contribution of buried distal amino acid residues in horse liver alcohol dehydrogenase to structure and catalysis.
Protein Sci., 27, 2018
|
|
5KJC
 
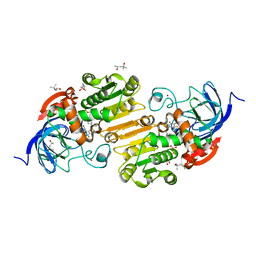 | | V222I horse liver alcohol dehydrogenase complexed with NAD+ and pentafluorobenzyl alcohol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V. | | Deposit date: | 2016-06-18 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Contribution of buried distal amino acid residues in horse liver alcohol dehydrogenase to structure and catalysis.
Protein Sci., 27, 2018
|
|
5KJE
 
 | | F322L horse liver alcohol dehydrogenase complexed with NAD+ and pentafluorobenzyl alcohol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V. | | Deposit date: | 2016-06-18 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Contribution of buried distal amino acid residues in horse liver alcohol dehydrogenase to structure and catalysis.
Protein Sci., 27, 2018
|
|
2G6F
 
 | |
1G9O
 
 | | FIRST PDZ DOMAIN OF THE HUMAN NA+/H+ EXCHANGER REGULATORY FACTOR | | Descriptor: | NHE-RF | | Authors: | Karthikeyan, S, Leung, T, Birrane, G, Webster, G, Ladias, J.A.A. | | Deposit date: | 2000-11-26 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the PDZ1 domain of human Na(+)/H(+) exchanger regulatory factor provides insights into the mechanism of carboxyl-terminal leucine recognition by class I PDZ domains.
J.Mol.Biol., 308, 2001
|
|
2ESE
 
 | | Structure of the SAM domain of Vts1p in complex with RNA | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*GP*GP*CP*UP*CP*UP*GP*GP*CP*AP*GP*CP*UP*UP*UP*UP*CP*C)-3', Vts1p | | Authors: | Allain, F.H.T. | | Deposit date: | 2005-10-26 | | Release date: | 2006-01-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Shape-specific recognition in the structure of the Vts1p SAM domain with RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2ES5
 
 | | Structure of the SRE RNA | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*GP*GP*CP*UP*CP*UP*GP*GP*CP*AP*GP*CP*UP*UP*UP*UP*CP*C)-3' | | Authors: | Allain, F.H.T. | | Deposit date: | 2005-10-25 | | Release date: | 2006-01-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Shape-specific recognition in the structure of the Vts1p SAM domain with RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
