4P9I
 
 | |
4P9L
 
 | |
4P9J
 
 | |
9FNR
 
 | | Artificial metalloenzyme with a nickel-based 1,10-phenanthroline cofactor and streptavidin S112V mutant | | Descriptor: | N-methyl-N-[(4,4,6,6-tetrahydroxy-4,6-dioxido-1,3,3a,5,6a-tetrahydrothien[3,4-d]imidazol-4-ium-2-yl)methyl]-5-(2,4,4-trihydroxy-2-keto-3,3a,5,6-tetrahydro-1H-thien[3,4-d]imidazol-4-ium-6-yl, SULFATE ION, Streptavidin | | Authors: | Lau, K, Wang, W, Pojer, F, Larabi, A. | | Deposit date: | 2024-06-10 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Artificial Metalloenzymes with Two Catalytic Cofactors for Tandem Abiotic Transformations.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9FOA
 
 | | Artificial photoenzyme with anthraquinone cofactor and wild type streptavidin | | Descriptor: | GLYCEROL, SULFATE ION, Streptavidin, ... | | Authors: | Lau, K, Wang, W, Pojer, F, Larabi, A. | | Deposit date: | 2024-06-11 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Artificial Metalloenzymes with Two Catalytic Cofactors for Tandem Abiotic Transformations.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8AQT
 
 | | Beta SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQU
 
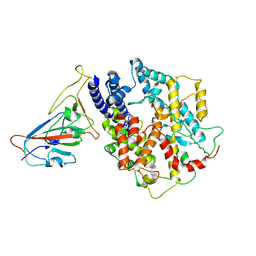 | | BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQV
 
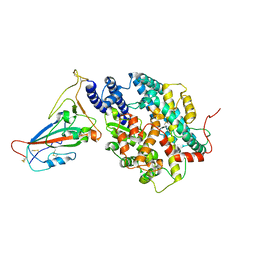 | | BA.2.12.1 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQS
 
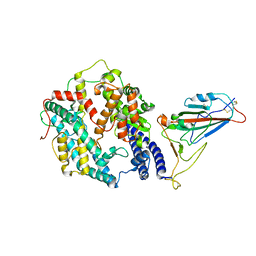 | | BA.4/5 SARS-CoV-2 Spike bound to human ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQW
 
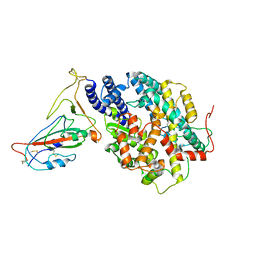 | | BA.4/5 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
9FFJ
 
 | | Artificial metalloenzyme with a nickel-based 1,10-phenanthroline cofactor and streptavidin N49M-S112V mutant | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lau, K, Wang, W, Pojer, F, Larabi, A. | | Deposit date: | 2024-05-23 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Artificial Metalloenzymes with Two Catalytic Cofactors for Tandem Abiotic Transformations.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9RDT
 
 | | Structure of the RhGB07 Prefusion, Closed-state trimeric spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RhGB07 Spike- T4 foldon fusion protein, ... | | Authors: | Lau, K, Dong, C.N, Ekundayo, B. | | Deposit date: | 2025-06-03 | | Release date: | 2025-09-24 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Understanding the future risk of bat coronavirus spillover into humans - correlating sarbecovirus receptor usage, host range, and antigenicity
Biorxiv, 2025
|
|
6Y4P
 
 | | Calmodulin N53I variant bound to cardiac ryanodine receptor (RyR2) calmodulin binding domain | | Descriptor: | CALCIUM ION, Calmodulin-1, Ryanodine receptor 2 | | Authors: | Lau, K, Nielsen, L.H, Holt, C, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Van Petegem, F, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13325572 Å) | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
6XZM
 
 | | Arabidopsis UV-B photoreceptor UVR8 mutant D96N D107N W285A | | Descriptor: | DI(HYDROXYETHYL)ETHER, NITRATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Lau, K, Hothorn, M. | | Deposit date: | 2020-02-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.10009313 Å) | | Cite: | A constitutively monomeric UVR8 photoreceptor confers enhanced UV-B photomorphogenesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Y4O
 
 | | Calmodulin bound to cardiac ryanodine receptor (RyR2) calmodulin binding domain | | Descriptor: | CALCIUM ION, Calmodulin-2, Ryanodine receptor 2 | | Authors: | Lau, K, Nielsen, L.H, Holt, C, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Van Petegem, F, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83549082 Å) | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
6XZN
 
 | | Arabidopsis UV-B photoreceptor UVR8 mutant G101S W285A | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, TRIETHYLENE GLYCOL, ... | | Authors: | Lau, K, Hothorn, M. | | Deposit date: | 2020-02-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A constitutively monomeric UVR8 photoreceptor confers enhanced UV-B photomorphogenesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6XZL
 
 | | Arabidopsis UV-B photoreceptor UVR8 mutant D96N D107N | | Descriptor: | GLYCEROL, Ultraviolet-B receptor UVR8 | | Authors: | Lau, K, Hothorn, M. | | Deposit date: | 2020-02-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A constitutively monomeric UVR8 photoreceptor confers enhanced UV-B photomorphogenesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6QTS
 
 | |
6QTR
 
 | |
6QTT
 
 | |
6QTV
 
 | |
6QTQ
 
 | |
6QTX
 
 | |
6QTO
 
 | |
6QTW
 
 | |
