2POS
 
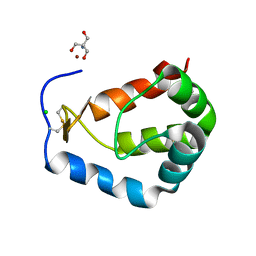 | | Crystal Structure of sylvaticin, a new secreted protein from pythium sylvaticum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Lascombe, M.B, Prange, T. | | Deposit date: | 2007-04-27 | | Release date: | 2008-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of sylvaticin, a new alpha-elicitin-like protein from Pythium sylvaticum.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2RKN
 
 | | X-ray structure of the self-defense and signaling protein DIR1 from Arabidopsis taliana | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, DIR1 protein, ZINC ION | | Authors: | Lascombe, M.B, Prange, T, Buhot, N, Marion, D, Bakan, B, Lamb, C. | | Deposit date: | 2007-10-17 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of "defective in induced resistance" protein of Arabidopsis thaliana, DIR1, reveals a new type of lipid transfer protein.
Protein Sci., 17, 2008
|
|
2PR0
 
 | | Crystal structure of Sylvaticin, a new secreted protein from Pythium Sylvaticum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NICKEL (II) ION, sylvaticin | | Authors: | Lascombe, M.B, Prange, T, Retailleau, P. | | Deposit date: | 2007-05-03 | | Release date: | 2008-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure of sylvaticin, a new alpha-elicitin-like protein from Pythium sylvaticum.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1FAI
 
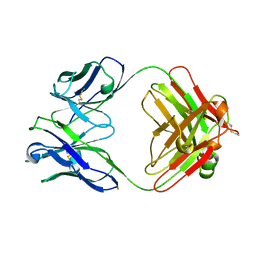 | | THREE-DIMENSIONAL STRUCTURE OF TWO CRYSTAL FORMS OF FAB R19.9, FROM A MONOCLONAL ANTI-ARSONATE ANTIBODY | | Descriptor: | IGG2B-KAPPA R19.9 FAB (HEAVY CHAIN), IGG2B-KAPPA R19.9 FAB (LIGHT CHAIN) | | Authors: | Lascombe, M.B, Alzari, P.M, Poljak, R.J, Nisonoff, A. | | Deposit date: | 1992-05-27 | | Release date: | 1992-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of two crystal forms of FabR19.9 from a monoclonal anti-arsonate antibody.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1EW3
 
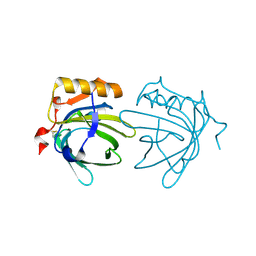 | | CRYSTAL STRUCTURE OF THE MAJOR HORSE ALLERGEN EQU C 1 | | Descriptor: | ALLERGEN EQU C 1 | | Authors: | Lascombe, M.B, Gregoire, C, Poncet, P, Tavares, G.A, Rosinski-Chupin, I, Rabillon, J, Goubran-Botros, H, Mazie, J.C, David, B, Alzari, P.M. | | Deposit date: | 2000-04-21 | | Release date: | 2000-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the allergen Equ c 1. A dimeric lipocalin with restricted IgE-reactive epitopes.
J.Biol.Chem., 275, 2000
|
|
2F19
 
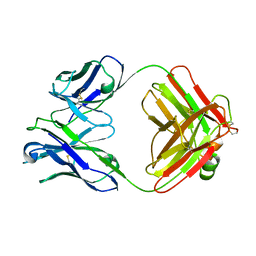 | | THREE-DIMENSIONAL STRUCTURE OF TWO CRYSTAL FORMS OF FAB R19.9, FROM A MONOCLONAL ANTI-ARSONATE ANTIBODY | | Descriptor: | IGG2B-KAPPA R19.9 FAB (HEAVY CHAIN), IGG2B-KAPPA R19.9 FAB (LIGHT CHAIN) | | Authors: | Lascombe, M.B, Alzari, P.M, Poljak, R.J, Nisonoff, A. | | Deposit date: | 1992-05-27 | | Release date: | 1992-10-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of two crystal forms of FabR19.9 from a monoclonal anti-arsonate antibody.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
3GSH
 
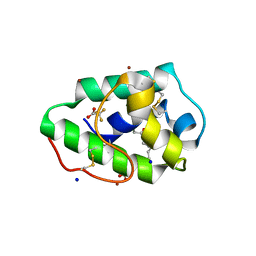 | | Three-dimensional structure of a post translational modified barley LTP1 | | Descriptor: | (12E)-10-oxooctadec-12-enoic acid, Non-specific lipid-transfer protein 1, SODIUM ION, ... | | Authors: | Lascombe, M.B, Prange, T, Bakan, B, Marion, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of oxylipin-conjugated barley LTP1 highlights the unique plasticity of the hydrophobic cavity of these plant lipid-binding proteins.
Biochem.Biophys.Res.Commun., 390, 2009
|
|
1CLC
 
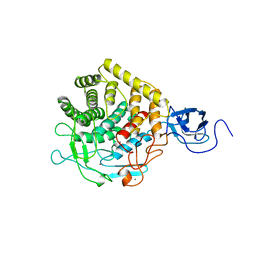 | |
6ZRE
 
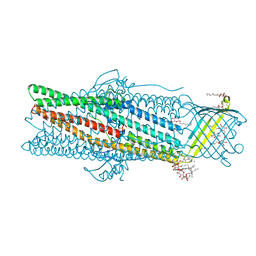 | | Deciphering the role of the channel constrictions in the opening mechanism of MexAB-OprM efflux pump from Pseudomonas aeruginosa | | Descriptor: | Outer membrane protein OprM, PALMITIC ACID, SULFATE ION, ... | | Authors: | Ntsogo Enguene, Y.V, Monlezun, L, Ma, M, Garnier, C, Lascombe, M.B, Salem, M, Guenard, S, Plesiat, P, Llanes, C, Phan, G, Broutin, I. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Deciphering the role of OprM constrictions in the opening mechanism of the MexAB-OprM efflux pump from Pseudomonas aeruginosa.
To Be Published
|
|
7AKZ
 
 | | Deciphering the role of the channel constrictions in the opening mechanism of MexAB-OprM efflux pump from Pseudomonas aeruginosa | | Descriptor: | Outer membrane protein OprM, PALMITIC ACID, octyl beta-D-glucopyranoside | | Authors: | Ntsogo Enguene, V.Y, Monlezun, L, Ma, M, Garnier, C, Lascombe, M.B, Salem, M, Guenard, S, Plesiat, P, Llanes, C, Phan, G, Broutin, I. | | Deposit date: | 2020-10-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Deciphering the role of the channel constrictions in the opening mechanism of MexAB-OprM efflux pump from Pseudomonas aeruginosa
To Be Published
|
|
1LRI
 
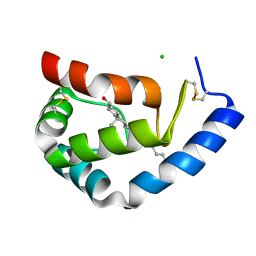 | | BETA-CRYPTOGEIN-CHOLESTEROL COMPLEX | | Descriptor: | Beta-elicitin cryptogein, CHLORIDE ION, CHOLESTEROL | | Authors: | Lascombe, M.-B, Ponchet, M, Venard, P, Milat, M.-L, Blein, J.-P, Prange, T. | | Deposit date: | 2002-05-15 | | Release date: | 2002-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The 1.45 A resolution structure of the cryptogein-cholesterol complex: a close-up view of a sterol carrier protein (SCP) active site.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KWF
 
 | | Atomic Resolution Structure of an Inverting Glycosidase in Complex with Substrate | | Descriptor: | Endoglucanase A, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Guerin, D.M.A, Lascombe, M.-B, Costabel, M, Souchon, H, Lamzin, V, Beguin, P, Alzari, P.M. | | Deposit date: | 2002-01-29 | | Release date: | 2002-03-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Atomic (0.94 A) resolution structure of an inverting glycosidase in complex with substrate.
J.Mol.Biol., 316, 2002
|
|
3D5K
 
 | |
4Y1K
 
 | |
