5OLM
 
 | | TRIM21 | | 分子名称: | E3 ubiquitin-protein ligase TRIM21, ZINC ION | | 著者 | James, L.C. | | 登録日 | 2017-07-28 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Intracellular antibody signalling is regulated by phosphorylation of the Fc receptor TRIM21.
Elife, 7, 2018
|
|
3H0O
 
 | | The importance of CH-Pi stacking interactions between carbohydrate and aromatic residues in truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-glucanase, ... | | 著者 | Tsai, L.C, Hsiao, C.H. | | 登録日 | 2009-04-09 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The importance of CH-Pi stacking interactions between carbohydrate and aromatic residues in truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase
To be Published
|
|
3HR9
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase F40I mutant | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-glucanase, ... | | 著者 | Tsai, L.C, Huang, H.C, Hsiao, C.H. | | 登録日 | 2009-06-09 | | 公開日 | 2009-07-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase mutant F40I
To be Published
|
|
6S53
 
 | | Crystal structure of TRIM21 RING domain in complex with an isopeptide-linked Ube2N~ubiquitin conjugate | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase TRIM21, Polyubiquitin-C, ... | | 著者 | Kiss, L, Boland, A, Neuhaus, D, James, L.C. | | 登録日 | 2019-06-30 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A tri-ionic anchor mechanism drives Ube2N-specific recruitment and K63-chain ubiquitination in TRIM ligases.
Nat Commun, 10, 2019
|
|
6RSS
 
 | | Solution structure of the fourth WW domain of WWP2 with GB1-tag | | 分子名称: | NEDD4-like E3 ubiquitin-protein ligase WWP2 | | 著者 | Wahl, L.C, Watt, J.E, Tolchard, J, Blumenschein, T.M.A, Chantry, A. | | 登録日 | 2019-05-22 | | 公開日 | 2019-10-09 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Smad7 Binds Differently to Individual and Tandem WW3 and WW4 Domains of WWP2 Ubiquitin Ligase Isoforms.
Int J Mol Sci, 20, 2019
|
|
7ZN5
 
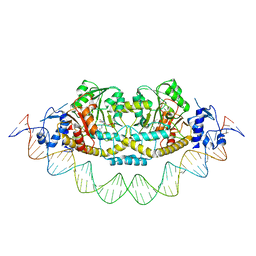 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry. | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | 登録日 | 2022-04-20 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZPA
 
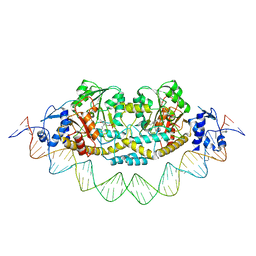 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | 登録日 | 2022-04-27 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZLA
 
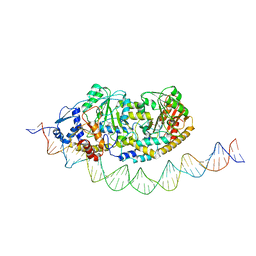 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | 登録日 | 2022-04-14 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.99 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZTH
 
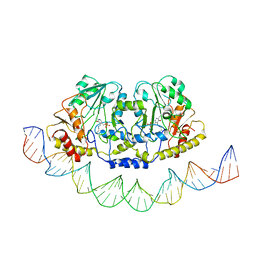 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | 登録日 | 2022-05-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
6TYW
 
 | |
6TYZ
 
 | |
6TYV
 
 | |
6TYX
 
 | |
6U7D
 
 | |
6TYU
 
 | |
6TYT
 
 | |
6VCJ
 
 | | Crystal structure of hsDHFR in complex with NADP+, DAP, and R-naproxen | | 分子名称: | (2R)-2-(6-methoxynaphthalen-2-yl)propanoic acid, Dihydrofolate reductase, FOLIC ACID, ... | | 著者 | Pedersen, L.C, London, R.E, Gabel, S.A, Krahn, J.M, DeRose, E.F. | | 登録日 | 2019-12-21 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | The Structural Basis for Nonsteroidal Anti-Inflammatory Drug Inhibition of Human Dihydrofolate Reductase.
J.Med.Chem., 63, 2020
|
|
2LZT
 
 | |
1FXD
 
 | | REFINED CRYSTAL STRUCTURE OF FERREDOXIN II FROM DESULFOVIBRIO GIGAS AT 1.7 ANGSTROMS | | 分子名称: | FE3-S4 CLUSTER, FERREDOXIN II | | 著者 | Kissinger, C.R, Sieker, L.C, Adman, E.T, Jensen, L.H. | | 登録日 | 1991-04-08 | | 公開日 | 1993-04-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Refined crystal structure of ferredoxin II from Desulfovibrio gigas at 1.7 A.
J.Mol.Biol., 219, 1991
|
|
6CDK
 
 | | Characterization of the P1+ intermediate state of nitrogenase P-cluster | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | 著者 | Keable, S.M, Zadvornyy, O.A, Rasmussen, A.J, Danyal, K, Eilers, B.J, Prussia, G.A, LeVan, A.X, Seefeldt, L.C, Peters, J.W. | | 登録日 | 2018-02-08 | | 公開日 | 2018-05-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural characterization of the P1+intermediate state of the P-cluster of nitrogenase.
J. Biol. Chem., 293, 2018
|
|
6D7N
 
 | |
4XE3
 
 | | OleP, the cytochrome P450 epoxidase from Streptomyces antibioticus involved in Oleandomycin biosynthesis: functional analysis and crystallographic structure in complex with clotrimazole. | | 分子名称: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Montemiglio, L.C, Parisi, G, Scaglione, A, Savino, C, Vallone, B. | | 登録日 | 2014-12-22 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Functional analysis and crystallographic structure of clotrimazole bound OleP, a cytochrome P450 epoxidase from Streptomyces antibioticus involved in oleandomycin biosynthesis.
Biochim.Biophys.Acta, 1860, 2015
|
|
6D7M
 
 | | Crystal structure of the W184R/W231R Importin alpha mutant | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Peroxidase,Importin subunit alpha-1, ... | | 著者 | Pedersen, L.C, London, R.E, Gabel, S.A. | | 登録日 | 2018-04-25 | | 公開日 | 2019-03-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.187 Å) | | 主引用文献 | Variations in nuclear localization strategies among pol X family enzymes.
Traffic, 2018
|
|
4XPI
 
 | | Fe protein independent substrate reduction by nitrogenase variants altered in intramolecular electron transfer | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, ... | | 著者 | Danyal, K, Rasmusen, A.J, Keable, S.M, Shaw, S, Zadvornyy, O, Duval, S, Dean, D.R, Raugei, S, Peters, J.W, Seefeldt, L.C. | | 登録日 | 2015-01-17 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Fe protein-independent substrate reduction by nitrogenase MoFe protein variants.
Biochemistry, 54, 2015
|
|
4XRO
 
 | |
