1EEQ
 
 | | M4L/Y(27D)D/T94H Mutant of LEN | | Descriptor: | KAPPA-4 IMMUNOGLOBULIN (LIGHT CHAIN) | | Authors: | Pokkuluri, P.R, Raffen, R, Dieckman, L, Boogaard, C, Stevens, F.J, Schiffer, M. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Increasing protein stability by polar surface residues: domain-wide consequences of interactions within a loop.
Biophys.J., 82, 2002
|
|
1IRK
 
 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF THE HUMAN INSULIN RECEPTOR | | Descriptor: | ETHYL MERCURY ION, INSULIN RECEPTOR TYROSINE KINASE DOMAIN | | Authors: | Hubbard, S.R, Wei, L, Ellis, L, Hendrickson, W.A. | | Deposit date: | 1995-01-02 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the tyrosine kinase domain of the human insulin receptor.
Nature, 372, 1994
|
|
6TQL
 
 | | Cryo-EM of elastase-treated human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
6TQK
 
 | | Cryo-EM of native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
1H15
 
 | | X-ray crystal structure of HLA-DRA1*0101/DRB5*0101 complexed with a peptide from Epstein Barr Virus DNA polymerase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA POLYMERASE, ... | | Authors: | Lang, H, Jacobsen, H, Ikemizu, S, Andersson, C, Harlos, K, Madsen, L, Hjorth, P, Sondergaard, L, Svejgaard, A, Wucherpfennig, K, Stuart, D.I, Bell, J.I, Jones, E.Y, Fugger, L. | | Deposit date: | 2002-07-02 | | Release date: | 2002-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Functional and Structural Basis for Tcr Cross-Reactivity in Multiple Sclerosis
Nat.Immunol., 3, 2002
|
|
4Y7N
 
 | | The Structure Insight into 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
4Y52
 
 | | Crystal structure of 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-11 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
8P7G
 
 | |
11BA
 
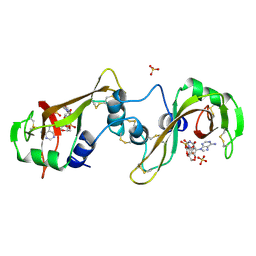 | | BINDING OF A SUBSTRATE ANALOGUE TO A DOMAIN SWAPPING PROTEIN IN THE COMPLEX OF BOVINE SEMINAL RIBONUCLEASE WITH URIDYLYL-2',5'-ADENOSINE | | Descriptor: | PROTEIN (RIBONUCLEASE, SEMINAL), SULFATE ION, ... | | Authors: | Vitagliano, L, Adinolfi, S, Riccio, A, Sica, F, Zagari, A, Mazzarella, L. | | Deposit date: | 1999-03-17 | | Release date: | 1999-03-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Binding of a substrate analog to a domain swapping protein: X-ray structure of the complex of bovine seminal ribonuclease with uridylyl(2',5')adenosine.
Protein Sci., 7, 1998
|
|
7PFP
 
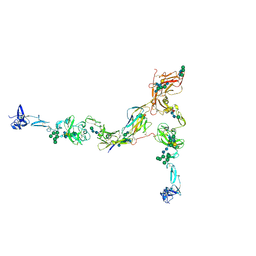 | | Full-length cryo-EM structure of the native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-08-11 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4TVC
 
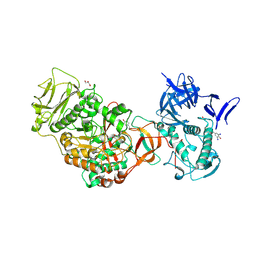 | | N-terminally truncated dextransucrase DSR-E from Leuconostoc mesenteroides NRRL B-1299 in complex with gluco-oligosaccharides | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL, ... | | Authors: | Brison, Y, Remaud-Simeon, M, Mourey, L, Tranier, S. | | Deposit date: | 2014-06-26 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into the Carbohydrate Binding Ability of an alpha-(12) Branching Sucrase from Glycoside Hydrolase Family 70.
J.Biol.Chem., 291, 2016
|
|
4TXM
 
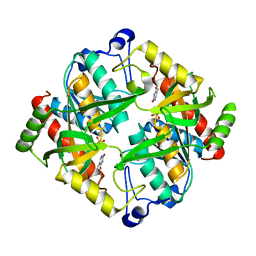 | | Crystal structure of uridine phosphorylase from Schistosoma mansoni in complex with thymine | | Descriptor: | SULFATE ION, THYMINE, Uridine phosphorylase | | Authors: | Marinho, A, Torini, J, Romanello, L, Cassago, A, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2014-07-03 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Analysis of two Schistosoma mansoni uridine phosphorylases isoforms suggests the emergence of a protein with a non-canonical function.
Biochimie, 125, 2016
|
|
4U1X
 
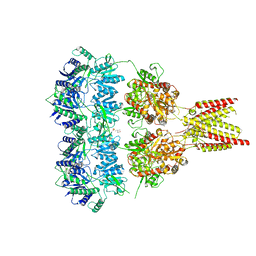 | | Full length GluA2-kainate-(R,R)-2b complex crystal form B | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4TVD
 
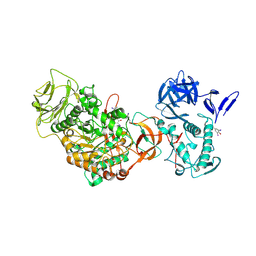 | | N-terminally truncated dextransucrase DSR-E from Leuconostoc mesenteroides NRRL B-1299 in complex with D-glucose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Dextransucrase, ... | | Authors: | Brison, Y, Remaud-Simeon, M, Mourey, L, Tranier, S. | | Deposit date: | 2014-06-26 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the Carbohydrate Binding Ability of an alpha-(12) Branching Sucrase from Glycoside Hydrolase Family 70.
J.Biol.Chem., 291, 2016
|
|
4TZ7
 
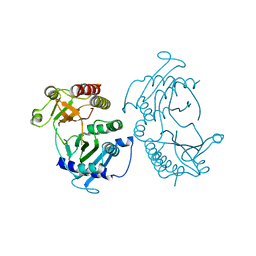 | | Crystal structure of type I phosphatidylinositol 4-phosphate 5-kinase alpha from Zebrafish | | Descriptor: | Phosphatidylinositol-4-phosphate 5-kinase, type I, alpha | | Authors: | Hu, J, Qin, Y, Wang, J, Li, L, Wu, D, Ha, Y. | | Deposit date: | 2014-07-09 | | Release date: | 2015-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Resolution of structure of PIP5K1A reveals molecular mechanism for its regulation by dimerization and dishevelled.
Nat Commun, 6, 2015
|
|
1EEH
 
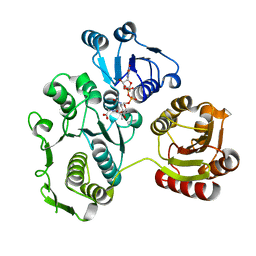 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE | | Authors: | Bertrand, J.A, Fanchon, E, Martin, L, Chantalat, L, Auger, G, Blanot, D, van Heijenoort, J, Dideberg, O. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | "Open" structures of MurD: domain movements and structural similarities with folylpolyglutamate synthetase.
J.Mol.Biol., 301, 2000
|
|
4U2Q
 
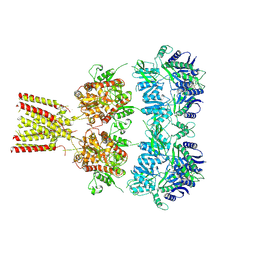 | | Full-length AMPA subtype ionotropic glutamate receptor GluA2 in complex with partial agonist kainate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Duerr, K.L, Chen, L, Gouaux, E. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5247 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U49
 
 | |
4U5J
 
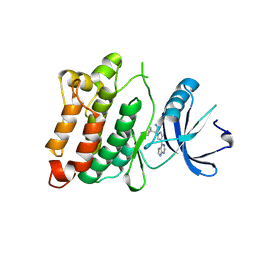 | | C-Src in complex with Ruxolitinib | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Chen, Y, Duan, Y, Chen, L. | | Deposit date: | 2014-07-25 | | Release date: | 2014-09-17 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | c-Src Binds to the Cancer Drug Ruxolitinib with an Active Conformation
Plos One, 9, 2014
|
|
4U2R
 
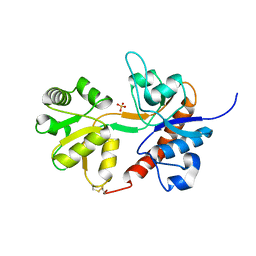 | | Crystal structure of the GLUR2 ligand binding core (S1S2J, flip variant) in the apo state | | Descriptor: | Glutamate receptor 2, SULFATE ION | | Authors: | Duerr, K.L, Chen, L, Gouaux, E. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4114 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U4B
 
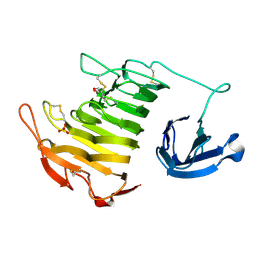 | |
4U1Z
 
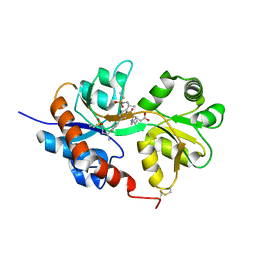 | | GluA2flip sLBD complexed with kainate and (R,R)-2b crystal form D | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9401 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U21
 
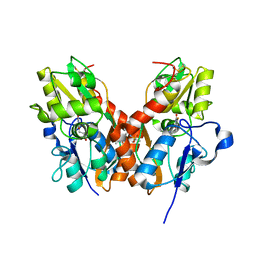 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form E | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3908 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U22
 
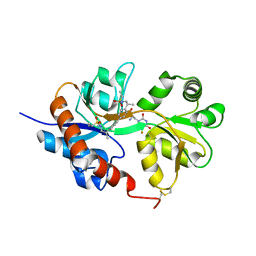 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form D | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4409 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U23
 
 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form F | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6734 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
