2YVP
 
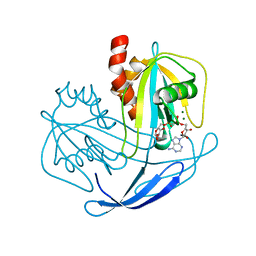 | | Crystal structure of NDX2 in complex with MG2+ and ampcpr from thermus thermophilus HB8 | | Descriptor: | MAGNESIUM ION, METHYLENE ADP-BETA-XYLOSE, MutT/nudix family protein | | Authors: | Wakamatsu, T, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-13 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis for different substrate specificities of two ADP-ribose pyrophosphatases from Thermus thermophilus HB8
J.Bacteriol., 190, 2008
|
|
2YV4
 
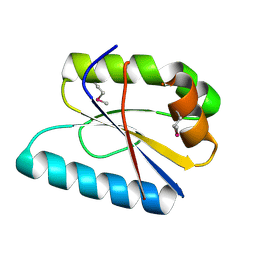 | |
2YVM
 
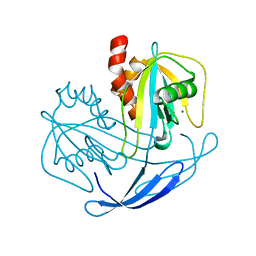 | | Crystal structure of NDX2 in complex with MG2+ from thermus thermophilus HB8 | | Descriptor: | MAGNESIUM ION, MutT/nudix family protein | | Authors: | Wakamatsu, T, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-13 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for different substrate specificities of two ADP-ribose pyrophosphatases from Thermus thermophilus HB8
J.Bacteriol., 190, 2008
|
|
2ZIE
 
 | | Crystal Structure of TTHA0409, Putatative DNA Modification Methylase from Thermus thermophilus HB8- Selenomethionine derivative | | Descriptor: | Putative modification methylase | | Authors: | Morita, R, Ishikawa, H, Nakagawa, N, Masui, R, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-02-15 | | Release date: | 2008-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative DNA methylase TTHA0409 from Thermus thermophilus HB8
Proteins, 73, 2008
|
|
2DKF
 
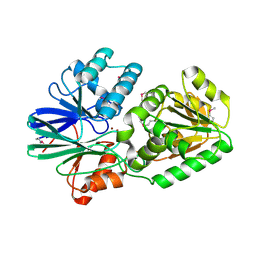 | | Crystal Structure of TTHA0252 from Thermus thermophilus HB8, a RNA Degradation Protein of the Metallo-beta-lactamase Superfamily | | Descriptor: | ZINC ION, metallo-beta-lactamase superfamily protein | | Authors: | Ishikawa, I, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-10 | | Release date: | 2006-12-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of TTHA0252 from Thermus thermophilus HB8, a RNA degradation protein of the metallo-beta-lactamase superfamily
J.Biochem.(Tokyo), 140, 2006
|
|
2ZIG
 
 | | Crystal Structure of TTHA0409, Putative DNA Modification Methylase from Thermus thermophilus HB8 | | Descriptor: | Putative modification methylase | | Authors: | Morita, R, Ishikawa, H, Nakagawa, N, Masui, R, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-02-15 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative DNA methylase TTHA0409 from Thermus thermophilus HB8
Proteins, 73, 2008
|
|
2ZIF
 
 | | Crystal Structure of TTHA0409, Putative DNA Modification Methylase from Thermus thermophilus HB8- Complexed with S-Adenosyl-L-Methionine | | Descriptor: | Putative modification methylase, S-ADENOSYLMETHIONINE | | Authors: | Morita, R, Ishikawa, H, Nakagawa, N, Masui, R, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-02-15 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative DNA methylase TTHA0409 from Thermus thermophilus HB8
Proteins, 73, 2008
|
|
3A0J
 
 | | Crystal structure of cold shock protein 1 from Thermus thermophilus HB8 | | Descriptor: | Cold shock protein | | Authors: | Miyazaki, T, Nakagawa, N, Kuramitsu, S, Masui, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-03-19 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Biological Action of Cold Shock Protein 1 from Thermus thermophilus HB8
To be Published
|
|
3A4Y
 
 | | Crystal Structure of H61A mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of H61A mutant TTHA0252 from Thermus thermophilus HB8
to be published
|
|
2YVS
 
 | |
2D64
 
 | | Aspartate Aminotransferase Mutant MABC With Isovaleric Acid | | Descriptor: | Aspartate aminotransferase, ISOVALERIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D61
 
 | | Aspartate Aminotransferase Mutant MA With Maleic Acid | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-08 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D63
 
 | | Aspartate Aminotransferase Mutant MA With Isovaleric Acid | | Descriptor: | Aspartate aminotransferase, ISOVALERIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D5Y
 
 | | Aspartate Aminotransferase Mutant MC With Isovaleric Acid | | Descriptor: | Aspartate aminotransferase, ISOVALERIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-08 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D7Y
 
 | | Aspartate Aminotransferase Mutant MA | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-30 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D3Y
 
 | | Crystal structure of uracil-DNA glycosylase from Thermus Thermophilus HB8 | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, ACETATE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
2D4R
 
 | | Crystal structure of TTHA0849 from Thermus thermophilus HB8 | | Descriptor: | SULFATE ION, hypothetical protein TTHA0849 | | Authors: | Nakabayashi, M, Shibata, N, Kuramitsu, S, Higuchi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-23 | | Release date: | 2005-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a conserved hypothetical protein, TTHA0849 from Thermus thermophilus HB8, at 2.4 A resolution: a putative member of the StAR-related lipid-transfer (START) domain superfamily.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2D65
 
 | | Aspartate Aminotransferase Mutant MABC | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D7Z
 
 | | Aspartate Aminotransferase Mutant MAB Complexed with Maleic Acid | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-30 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D66
 
 | | Aspartate Aminotransferase Mutant MAB | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2DDG
 
 | | Crystal structure of uracil-DNA glycosylase in complex with AP:G containing DNA | | Descriptor: | 5'-D(*AP*TP*GP*TP*TP*GP*CP*(D1P)P*TP*TP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*AP*AP*GP*GP*CP*AP*AP*CP*A)-3', ACETATE ION, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Hoseki, J, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-28 | | Release date: | 2007-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
2DP6
 
 | | Crystal structure of uracil-DNA glycosylase in complex with AP:C containing DNA | | Descriptor: | 5'-D(*AP*TP*GP*TP*TP*GP*CP*(D1P)P*TP*TP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*AP*AP*CP*GP*CP*AP*AP*CP*A)-3', DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Kuramitsu, S, Hoseki, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-07 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Family 5 Uracil-DNA Glycosylase Bound to DNA Reveals Insights into the Mechanism for Substrate Recognition and Catalysis
To be Published
|
|
2ZQE
 
 | | Crystal structure of the Smr domain of Thermus thermophilus MutS2 | | Descriptor: | MutS2 protein | | Authors: | Fukui, K, Kitamura, Y, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2008-08-08 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MutS2 endonuclease domain and the mechanism of homologous recombination suppression
J.Biol.Chem., 283, 2008
|
|
2DEM
 
 | | Crystal structure of Uracil-DNA glycosylase in complex with AP:A containing DNA | | Descriptor: | 5'-D(*AP*TP*GP*TP*TP*GP*CP*(D1P)P*TP*TP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*AP*AP*AP*GP*CP*AP*AP*CP*A)-3', DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Hoseki, J, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
2ZKI
 
 | | Crystal structure of hypothetical Trp repressor binding protein from Sul folobus tokodaii (ST0872) | | Descriptor: | 199aa long hypothetical Trp repressor binding protein, SULFATE ION | | Authors: | Kawano, T, Teshima, N, Suzuki, A, Kuramitsu, S, Yamane, T. | | Deposit date: | 2008-03-21 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of hypothetical Trp repressor binding protein from Sul
folobus tokodaii (ST0872)
To be Published
|
|
