3BWI
 
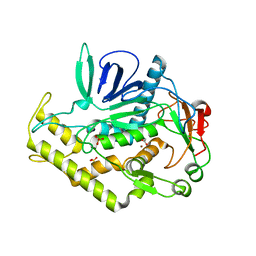 | | Crystal structure of the catalytic domain of botulinum neurotoxin serotype A with an acetate ion bound at the active site | | 分子名称: | ACETATE ION, Botulinum neurotoxin A light chain, SULFATE ION, ... | | 著者 | Kumaran, D, Rawat, R, Swaminathan, S. | | 登録日 | 2008-01-09 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure- and Substrate-based Inhibitor Design for Clostridium botulinum Neurotoxin Serotype A
J.Biol.Chem., 283, 2008
|
|
3C88
 
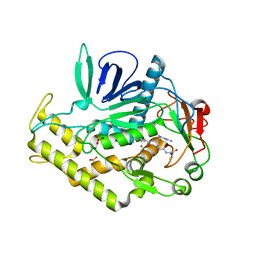 | |
3C8B
 
 | |
3C89
 
 | |
3CIH
 
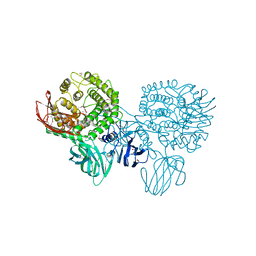 | | Crystal structure of a putative alpha-rhamnosidase from Bacteroides thetaiotaomicron | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative alpha-rhamnosidase | | 著者 | Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-03-11 | | 公開日 | 2008-04-01 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Crystal structure of a putative alpha-rhamnosidase from Bacteroides thetaiotaomicron.
To be Published
|
|
2AFA
 
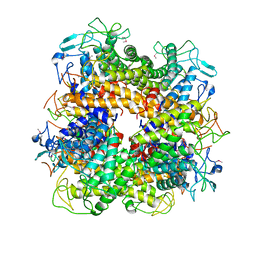 | |
4DUP
 
 | | Crystal Structure of a quinone oxidoreductase from Rhizobium etli CFN 42 | | 分子名称: | quinone oxidoreductase | | 著者 | Kumaran, D, Rice, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-02-22 | | 公開日 | 2012-03-07 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal Structure of a quinone oxidoreductase from Rhizobium etli CFN 42
To be Published
|
|
4E3Z
 
 | | Crystal Structure of a oxidoreductase from Rhizobium etli CFN 42 | | 分子名称: | Putative oxidoreductase protein | | 著者 | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-03-11 | | 公開日 | 2012-03-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of a oxidoreductase from Rhizobium etli CFN 42
To be Published
|
|
4QB5
 
 | | Crystal structure of a glyoxalase/bleomycin resistance protein from Albidiferax ferrireducens T118 | | 分子名称: | 1,2-ETHANEDIOL, Glyoxalase/bleomycin resistance protein/dioxygenase, SULFATE ION | | 著者 | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-05-06 | | 公開日 | 2014-07-23 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of a glyoxalase/bleomycin resistance protein from Albidiferax ferrireducens T118
To be Published
|
|
3US8
 
 | | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021 | | 分子名称: | Isocitrate dehydrogenase [NADP], SULFATE ION | | 著者 | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-11-23 | | 公開日 | 2011-12-14 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
3URH
 
 | | Crystal structure of a dihydrolipoamide dehydrogenase from Sinorhizobium meliloti 1021 | | 分子名称: | 1,2-ETHANEDIOL, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-11-22 | | 公開日 | 2011-12-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of a dihydrolipoamide dehydrogenase from Sinorhizobium meliloti 1021
TO BE PUBLISHED
|
|
3V8B
 
 | | Crystal Structure of a 3-ketoacyl-ACP reductase from Sinorhizobium meliloti 1021 | | 分子名称: | Putative dehydrogenase, possibly 3-oxoacyl-[acyl-carrier protein] reductase | | 著者 | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-12-22 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structure of a 3-ketoacyl-ACP reductase from Sinorhizobium meliloti 1021
To be Published
|
|
3BBL
 
 | |
3BGA
 
 | |
3C8A
 
 | |
1NJR
 
 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase | | 分子名称: | 32.1 kDa protein in ADH3-RCA1 intergenic region, Xylitol | | 著者 | Kumaran, D, Eswaramoorthy, S, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2003-01-02 | | 公開日 | 2004-08-17 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and mechanism of ADP-ribose-1''-monophosphatase (Appr-1''-pase), a ubiquitous cellular processing enzyme
Protein Sci., 14, 2005
|
|
3OC4
 
 | | Crystal Structure of a pyridine nucleotide-disulfide family oxidoreductase from the Enterococcus faecalis V583 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase, pyridine nucleotide-disulfide family, ... | | 著者 | Kumaran, D, Baumann, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-08-09 | | 公開日 | 2010-10-06 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of a pyridine nucleotide-disulfide family oxidoreductase from the Enterococcus faecalis V583
To be Published
|
|
1I4P
 
 | |
4ZJX
 
 | |
1I4R
 
 | |
1I4Q
 
 | |
3EUW
 
 | |
1F89
 
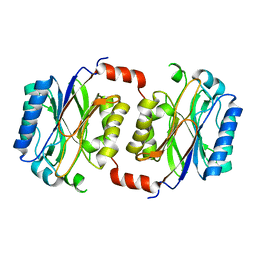 | | Crystal structure of Saccharomyces cerevisiae Nit3, a member of branch 10 of the nitrilase superfamily | | 分子名称: | 32.5 KDA PROTEIN YLR351C | | 著者 | Kumaran, D, Eswaramoorthy, S, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2000-06-29 | | 公開日 | 2001-10-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of a putative CN hydrolase from yeast
Proteins, 52, 2003
|
|
3FFZ
 
 | | Domain organization in Clostridium butulinum neurotoxin type E is unique: Its implication in faster translocation | | 分子名称: | ACETATE ION, Botulinum neurotoxin type E, SODIUM ION, ... | | 著者 | Kumaran, D, Eswaramoorthy, S, Swaminathan, S. | | 登録日 | 2008-12-04 | | 公開日 | 2008-12-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Domain organization in Clostridium botulinum neurotoxin type E is unique: its implication in faster translocation.
J.Mol.Biol., 386, 2009
|
|
1F1M
 
 | | CRYSTAL STRUCTURE OF OUTER SURFACE PROTEIN C (OSPC) | | 分子名称: | OUTER SURFACE PROTEIN C, ZINC ION | | 著者 | Kumaran, D, Eswaramoorthy, S, Dunn, J.J, Swaminathan, S. | | 登録日 | 2000-05-19 | | 公開日 | 2001-04-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of outer surface protein C (OspC) from the Lyme disease spirochete, Borrelia burgdorferi.
EMBO J., 20, 2001
|
|
