3ZC9
 
 | | Crystal Structure of Murraya koenigii Miraculin-Like Protein at 2.2 A resolution at pH 4.6 | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Selvakumar, P, Sharma, N, Tomar, P.P.S, Kumar, P, Sharma, A.K. | | Deposit date: | 2012-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Insights Into the Aggregation Behavior of Murraya Koenigii Miraculin-Like Protein Below Ph 7.5.
Proteins, 82, 2014
|
|
3ZC8
 
 | | Crystal Structure of Murraya koenigii Miraculin-Like Protein at 2.2 A resolution at pH 7.0 | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Selvakumar, P, Sharma, N, Tomar, P.P.S, Kumar, P, Sharma, A.K. | | Deposit date: | 2012-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Insights Into the Aggregation Behavior of Murraya Koenigii Miraculin-Like Protein Below Ph 7.5.
Proteins, 82, 2014
|
|
6AAL
 
 | | Crystal Structure of putative amino acid binding periplasmic ABC transporter protein from Candidatus Liberibacter asiaticus in complex with Arginine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ARGININE, ... | | Authors: | Kumar, P, Kesari, P, Ghosh, D.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-07-18 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of a putative periplasmic cystine-binding protein from Candidatus Liberibacter asiaticus: insights into an adapted mechanism of ligand binding.
Febs J., 286, 2019
|
|
2XSH
 
 | | CRYSTAL STRUCTURE OF P4 VARIANT OF BIPHENYL DIOXYGENASE FROM BURKHOLDERIA XENOVORANS LB400 IN COMPLEX WITH 2,6 DI CHLOROBIPHENYL | | Descriptor: | 2,6-DICHLOROBIPHENYL, BIPHENYL DIOXYGENASE SUBUNIT ALPHA, BIPHENYL DIOXYGENASE SUBUNIT BETA, ... | | Authors: | Kumar, P, Bolin, J.T. | | Deposit date: | 2010-09-29 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Insight Into the Expanded Pcb-Degrading Abilities of a Biphenyl Dioxygenase Obtained by Directed Evolution.
J.Mol.Biol., 405, 2011
|
|
2XSO
 
 | |
2XR8
 
 | |
2XRX
 
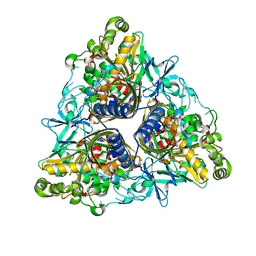 | |
2YFL
 
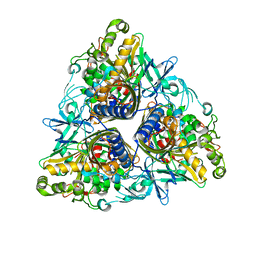 | |
5E3C
 
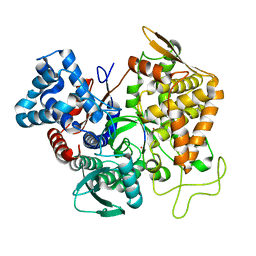 | | Structure of human DPP3 in complex with hemorphin like opioid peptide IVYPW | | Descriptor: | Dipeptidyl peptidase 3, IVYPW, MAGNESIUM ION, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.765 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
6O5U
 
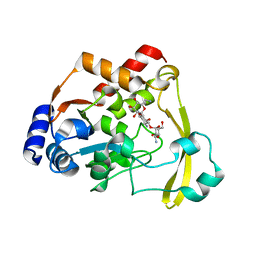 | | AAC-VIa bound to Kanamycin A | | Descriptor: | Aminoglycoside N(3)-acetyltransferase, KANAMYCIN A, MAGNESIUM ION | | Authors: | Kumar, P, Cuneo, M.J. | | Deposit date: | 2019-03-04 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5EGY
 
 | | Structure of ligand free human DPP3 in closed form. | | Descriptor: | Dipeptidyl peptidase 3, MAGNESIUM ION, ZINC ION | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.741 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
5EHH
 
 | | Structure of human DPP3 in complex with endomorphin-2. | | Descriptor: | Dipeptidyl peptidase 3, Endomorphin-2, MAGNESIUM ION, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-28 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
5E3A
 
 | | Structure of human DPP3 in complex with opioid peptide leu-enkephalin | | Descriptor: | Dipeptidyl peptidase 3, Leu-enkephalin, MAGNESIUM ION, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
4R62
 
 | | Structure of Rad6~Ub | | Descriptor: | ACETATE ION, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 2 | | Authors: | Kumar, P, Wolberger, C. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Role of a non-canonical surface of Rad6 in ubiquitin conjugating activity.
Nucleic Acids Res., 43, 2015
|
|
5UWV
 
 | | Crystal structure of Mycobacterium abscessus L,D-transpeptidase 2 | | Descriptor: | L,D-TRANSPEPTIDASE 2 | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2017-02-21 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Mycobacterium abscessus l,d-Transpeptidases Are Susceptible to Inactivation by Carbapenems and Cephalosporins but Not Penicillins.
Antimicrob. Agents Chemother., 61, 2017
|
|
7QDI
 
 | | Structure of octameric left-handed 310-helix bundle: D-310HD | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2021-11-27 | | Release date: | 2022-04-27 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
7QDJ
 
 | | Racemic structure of PK-10 and PK-11 | | Descriptor: | GLYCEROL, MALONATE ION, PK-10+PK-11, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2021-11-27 | | Release date: | 2022-04-27 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
7QDK
 
 | | A trimeric de novo coiled-coil assembly: CC-TypeN-LaLd | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CC-TypeN-LaLd, GLYCEROL, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2021-11-27 | | Release date: | 2022-04-27 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
8BFD
 
 | | Racemic structure of PK-7 (310HD-U2U5) | | Descriptor: | 310HD-U2U5, D-310HD-U2U5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
8BFE
 
 | | A dimeric de novo coiled-coil assembly: PK-2 (CC-TypeN-LaUbUcLd) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CC-TypeN-LaUbUcLd, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
8B45
 
 | | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4 | | Descriptor: | 1,2-ETHANEDIOL, CC-Tri-(UbUc)4, SODIUM ION, ... | | Authors: | Kumar, P, Martin, F.J.O, Dawson, W.M, Zieleniewski, F, Woolfson, D.N. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4
To Be Published
|
|
5E1G
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T208 | | Descriptor: | (2~{S},3~{R},4~{R})-4-(2-cyclohexyloxy-2-oxidanylidene-ethyl)sulfanyl-3-methyl-2-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, L,D-transpeptidase 2 | | Authors: | Kumar, P, Lamichhane, G, Ginell, S.L. | | Deposit date: | 2015-09-29 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5DZP
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T206 in conformation B | | Descriptor: | (2~{R},3~{R},4~{R})-4-methyl-3-(2-oxidanylidene-2-propoxy-ethyl)sulfanyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, L,D-transpeptidase 2 | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
6NP4
 
 | | AAC-VIa bound to Tobramycin | | Descriptor: | Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION, TOBRAMYCIN | | Authors: | Kumar, P, Cuneo, M.J. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6NP1
 
 | |
