6VBG
 
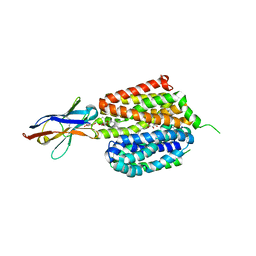 | | Lactose permease complex with thiodigalactoside and nanobody 9043 | | Descriptor: | Galactoside permease, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose, nanobody 9043, ... | | Authors: | Kumar, H, Stroud, R.M, Kaback, H.R, Finer-Moore, J, Smirnova, I, Kasho, V, Pardon, E, Steyart, J. | | Deposit date: | 2019-12-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Diversity in kinetics correlated with structure in nano body-stabilized LacY.
Plos One, 15, 2020
|
|
5N2I
 
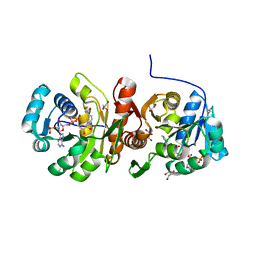 | | F420:NADPH oxidoreductase from Thermobifida fusca with NADP+ bound | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Reduced coenzyme F420:NADP oxidoreductase | | Authors: | Kumar, H, Nguyen, Q.-T, Binda, C, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2017-02-07 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isolation and characterization of a thermostable F420:NADPH oxidoreductase from Thermobifida fusca.
J. Biol. Chem., 292, 2017
|
|
6C9W
 
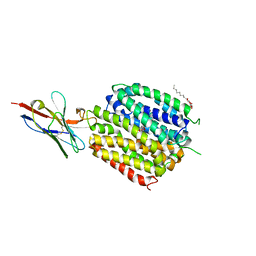 | | Crystal Structure of a ligand bound LacY/Nanobody Complex | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Lactose permease, Nanobody9047, ... | | Authors: | Kumar, H, Finer-Moore, J.S, Jiang, X, Smirnova, I, Kasho, V, Pardon, E, Steyaert, J, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a ligand-bound LacY-Nanobody Complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4ZYR
 
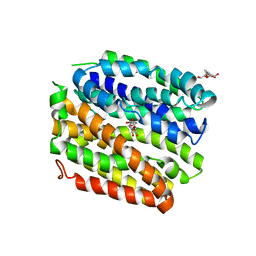 | | Crystal structure of E. coli Lactose permease G46W/G262W bound to p-nitrophenyl alpha-D-galactopyranoside (alpha-NPG) | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Lactose permease, nonyl beta-D-glucopyranoside | | Authors: | Kumar, H, Finer-Moore, J.S, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.312 Å) | | Cite: | Structure of LacY with an alpha-substituted galactoside: Connecting the binding site to the protonation site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8ER1
 
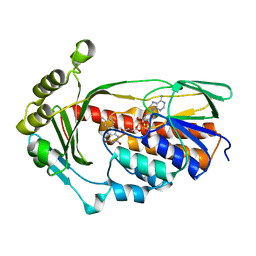 | | X-ray crystal structure of Tet(X6) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase | | Authors: | Kumar, H, Tolia, N.H. | | Deposit date: | 2022-10-11 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of anhydrotetracycline-bound Tet(X6) reveals the mechanism for inhibition of type 1 tetracycline destructases.
Commun Biol, 6, 2023
|
|
8ER0
 
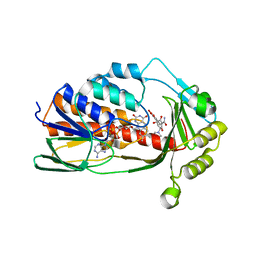 | | X-ray crystal structure of Tet(X6) bound to anhydrotetracycline | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase | | Authors: | Kumar, H, Tolia, N.H. | | Deposit date: | 2022-10-11 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of anhydrotetracycline-bound Tet(X6) reveals the mechanism for inhibition of type 1 tetracycline destructases.
Commun Biol, 6, 2023
|
|
4OAA
 
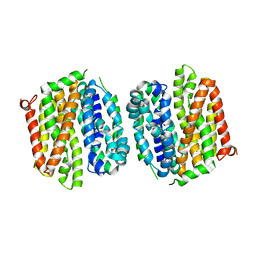 | | Crystal structure of E. coli lactose permease G46W,G262W bound to sugar | | Descriptor: | Lactose/galactose transporter, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose | | Authors: | Kumar, H, Kasho, V, Smirnova, I, Finer-Moore, J, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2014-01-03 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of sugar-bound LacY.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8TWG
 
 | |
8TWF
 
 | |
6WG9
 
 | | Crystal structure of tetracycline destructase Tet(X7) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Tetracycline destructase Tet(X7) | | Authors: | Kumar, H, Tolia, N.H. | | Deposit date: | 2020-04-05 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Tetracycline-inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad-spectrum tetracycline resistance.
Commun Biol, 3, 2020
|
|
7SP5
 
 | | Crystal Structure of a Eukaryotic Phosphate Transporter | | Descriptor: | PHOSPHATE ION, Phosphate transporter, nonyl beta-D-glucopyranoside | | Authors: | Stroud, R.M, Pedersen, B.P, Kumar, H, Waight, A.B, Risenmay, A.J, Roe-Zurz, Z, Chau, B.H, Schlessinger, A, Bonomi, M, Harries, W, Sali, A, Johri, A.K, Finer-Moore, J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a eukaryotic phosphate transporter.
Nature, 496, 2013
|
|
8FVZ
 
 | | PiPT Y150A | | Descriptor: | CITRATE ANION, PHOSPHATE ION, Phosphate transporter | | Authors: | Gupta, M, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2023-01-20 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Roles of PiPT residues in phosphate binding and transport tested by mutagenesis
To be published
|
|
8UKH
 
 | |
8ULF
 
 | |
