3PC4
 
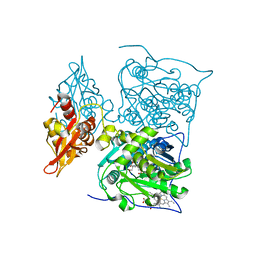 | |
3PC2
 
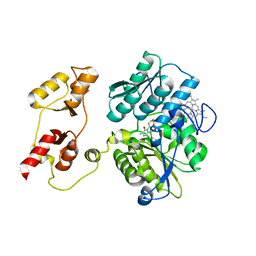 | |
3PC3
 
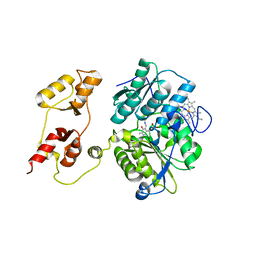 | |
3SBY
 
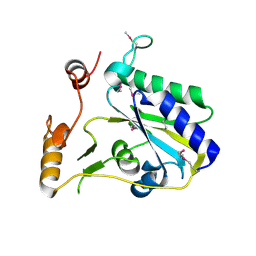 | | Crystal Structure of SeMet-Substituted Apo-MMACHC (1-244), a human B12 processing enzyme | | Descriptor: | Methylmalonic aciduria and homocystinuria type C protein | | Authors: | Koutmos, M, Gherasim, C, Smith, J.L, Banerjee, R. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural basis of multifunctionality in a vitamin B12-processing enzyme.
J.Biol.Chem., 286, 2011
|
|
3SC0
 
 | | Crystal Structure of MMACHC (1-238), a human B12 processing enzyme, complexed with MethylCobalamin | | Descriptor: | CO-METHYLCOBALAMIN, Methylmalonic aciduria and homocystinuria type C protein | | Authors: | Koutmos, M, Gherasim, C, Smith, J.L, Banerjee, R. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of multifunctionality in a vitamin B12-processing enzyme.
J.Biol.Chem., 286, 2011
|
|
3SBZ
 
 | | Crystal Structure of Apo-MMACHC (1-244), a human B12 processing enzyme | | Descriptor: | GLYCEROL, MALONATE ION, Methylmalonic aciduria and homocystinuria type C protein | | Authors: | Koutmos, M, Gherasim, C, Smith, J.L, Banerjee, R. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of multifunctionality in a vitamin B12-processing enzyme.
J.Biol.Chem., 286, 2011
|
|
3BOF
 
 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima complexed with Zn2+ and Homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, POTASSIUM ION, ... | | Authors: | Koutmos, M, Smith, J.L, Ludwig, M.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal active site elasticity linked to activation of homocysteine in methionine synthases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BOL
 
 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima complexed with Zn2+ | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, POTASSIUM ION, ... | | Authors: | Koutmos, M, Smith, J.L, Ludwig, M.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Metal active site elasticity linked to activation of homocysteine in methionine synthases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4G25
 
 | | Crystal Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana, SeMet substituted form with Sr | | Descriptor: | Pentatricopeptide repeat-containing protein At2g32230, mitochondrial, STRONTIUM ION, ... | | Authors: | Koutmos, M, Howard, M.J, Fierke, C.A. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mitochondrial ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5' processing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G23
 
 | |
4G26
 
 | | Crystal Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana with Ca | | Descriptor: | CALCIUM ION, Pentatricopeptide repeat-containing protein At2g32230, mitochondrial, ... | | Authors: | Koutmos, M, Howard, M.J, Fierke, C.A. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mitochondrial ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5' processing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G24
 
 | | Crystal Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana with Mn | | Descriptor: | 6-AMINOHEXANOIC ACID, MANGANESE (II) ION, Pentatricopeptide repeat-containing protein At2g32230, ... | | Authors: | Koutmos, M, Howard, M.J, Fierke, C.A. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mitochondrial ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5' processing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4LC1
 
 | | MeaB, A Bacterial Homolog of MMAA, Bound to GDP and crystallized in the presence of GDP and [AlF4]- | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Methylmalonyl-CoA mutase accessory protein | | Authors: | Koutmos, M, Padovani, D, Lofgren, M, Banerjee, R. | | Deposit date: | 2013-06-21 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Autoinhibition and Signaling by the Switch II Motif in the G-protein Chaperone of a Radical B12 Enzyme.
J.Biol.Chem., 288, 2013
|
|
3IV9
 
 | |
4M3P
 
 | | Betaine-Homocysteine S-Methyltransferase from Homo sapiens complexed with Homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Betaine--homocysteine S-methyltransferase 1, POTASSIUM ION, ... | | Authors: | Koutmos, M, Yamada, K, Mladkova, J, Paterova, J, Diamond, C.E, Tryon, K, Jungwirth, P, Garrow, T.A, Jiracek, J. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Specific potassium ion interactions facilitate homocysteine binding to betaine-homocysteine S-methyltransferase.
Proteins, 82, 2014
|
|
3IVA
 
 | | Structure of the B12-dependent Methionine Synthase (MetH) C-teminal half with AdoHcy bound | | Descriptor: | COBALAMIN, Methionine synthase, NITRATE ION, ... | | Authors: | Pattridge, K.A, Koutmos, M, Smith, J.L. | | Deposit date: | 2009-08-31 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the reactivation of cobalamin-dependent methionine synthase.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4JYC
 
 | | MeaB, A Bacterial Homolog of MMAA, in its Apo form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Methylmalonyl-CoA mutase accessory protein | | Authors: | Koutmos, M, Lofgren, M, Padovani, D, Banerjee, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A switch III motif relays signaling between a B12 enzyme and its G-protein chaperone.
Nat.Chem.Biol., 9, 2013
|
|
4JYB
 
 | | MeaB, A Bacterial Homolog of MMAA, Bound to GMPPNP | | Descriptor: | Methylmalonyl-CoA mutase accessory protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Koutmos, M, Lofgren, M, Padovani, D, Banerjee, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A switch III motif relays signaling between a B12 enzyme and its G-protein chaperone.
Nat.Chem.Biol., 9, 2013
|
|
4JGT
 
 | | Structure and kinetic analysis of H2S production by human Mercaptopyruvate Sulfurtransferase | | Descriptor: | 3-mercaptopyruvate sulfurtransferase, GLYCEROL, PYRUVIC ACID, ... | | Authors: | Koutmos, M, Yamada, K, Yadav, P.K, Chiku, T, Banerjee, R. | | Deposit date: | 2013-03-03 | | Release date: | 2013-05-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.161 Å) | | Cite: | Structure and Kinetic Analysis of H2S Production by Human Mercaptopyruvate Sulfurtransferase.
J.Biol.Chem., 288, 2013
|
|
5VOO
 
 | | Methionine synthase folate-binding domain with methyltetrahydrofolate from Thermus thermophilus HB8 | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydrofolate homocysteine S-methyltransferase, CHLORIDE ION, ... | | Authors: | Koutmos, M, Yamada, K. | | Deposit date: | 2017-05-03 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The folate-binding module of Thermus thermophilus cobalamin-dependent methionine synthase displays a distinct variation of the classical TIM barrel: a TIM barrel with a `twist'.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5VOP
 
 | | Methionine synthase folate-binding domain from Thermus thermophilus HB8 native | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydrofolate homocysteine S-methyltransferase, CITRATE ANION, ... | | Authors: | Koutmos, M, Yamada, K. | | Deposit date: | 2017-05-03 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The folate-binding module of Thermus thermophilus cobalamin-dependent methionine synthase displays a distinct variation of the classical TIM barrel: a TIM barrel with a `twist'.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
3BUL
 
 | |
6X8Z
 
 | | Crystal structure of N-truncated human B12 chaperone CblD(C262S)-thiolato-cob(III)alamin complex (108-296) | | Descriptor: | COBALAMIN, Methylmalonic aciduria and homocystinuria type D protein, mitochondrial | | Authors: | Mascarenhas, R, Li, Z, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Interprotein Co-S Coordination Complex in the B 12 -Trafficking Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6OXD
 
 | | Structure of Mycobacterium tuberculosis methylmalonyl-CoA mutase with adenosyl cobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Itaconyl coenzyme A, ... | | Authors: | Purchal, M, Ruetz, M, Banerjee, R, Koutmos, M. | | Deposit date: | 2019-05-13 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Itaconyl-CoA forms a stable biradical in methylmalonyl-CoA mutase and derails its activity and repair.
Science, 366, 2019
|
|
6OXC
 
 | | Structure of Mycobacterium tuberculosis methylmalonyl-CoA mutase with adenosyl cobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Methylmalonyl-CoA mutase large subunit, ... | | Authors: | Purchal, M, Ruetz, M, Banerjee, R, Koutmos, M. | | Deposit date: | 2019-05-13 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Itaconyl-CoA forms a stable biradical in methylmalonyl-CoA mutase and derails its activity and repair.
Science, 366, 2019
|
|
