1QFX
 
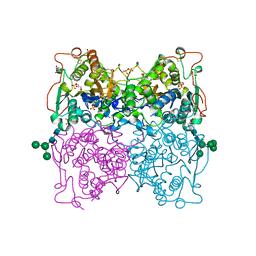 | | PH 2.5 ACID PHOSPHATASE FROM ASPERGILLUS NIGER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PROTEIN (PH 2.5 ACID PHOSPHATASE), ... | | Authors: | Kostrewa, D, Wyss, M, D'Arcy, A, Van Loon, A.P.G.M. | | Deposit date: | 1999-04-15 | | Release date: | 2000-04-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Aspergillus niger pH 2.5 acid phosphatase at 2. 4 A resolution.
J.Mol.Biol., 288, 1999
|
|
1FIA
 
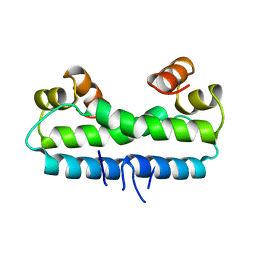 | | CRYSTAL STRUCTURE OF THE FACTOR FOR INVERSION STIMULATION FIS AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Kostrewa, D, Granzin, J, Choe, H.-W, Labahn, J, Saenger, W. | | Deposit date: | 1991-12-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the factor for inversion stimulation FIS at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
3K1F
 
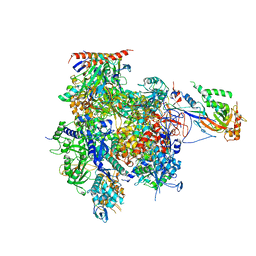 | | Crystal structure of RNA Polymerase II in complex with TFIIB | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Kostrewa, D, Zeller, M.E, Armache, K.-J, Seizl, M, Leike, K, Thomm, M, Cramer, P. | | Deposit date: | 2009-09-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | RNA polymerase II-TFIIB structure and mechanism of transcription initiation.
Nature, 462, 2009
|
|
1IHP
 
 | | STRUCTURE OF PHOSPHOMONOESTERASE | | Descriptor: | PHYTASE, SULFATE ION | | Authors: | Kostrewa, D. | | Deposit date: | 1997-02-04 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of phytase from Aspergillus ficuum at 2.5 A resolution.
Nat.Struct.Biol., 4, 1997
|
|
4YM7
 
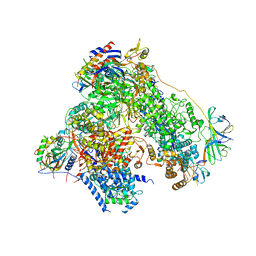 | | RNA polymerase I structure with an alternative dimer hinge | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Kostrewa, D, Kuhn, C.-D, Engel, C, Cramer, P. | | Deposit date: | 2015-03-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | An alternative RNA polymerase I structure reveals a dimer hinge.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3RNT
 
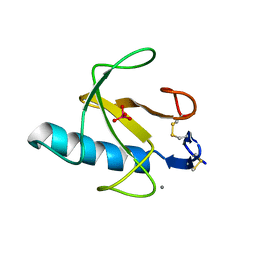 | | CRYSTAL STRUCTURE OF GUANOSINE-FREE RIBONUCLEASE T1, COMPLEXED WITH VANADATE(V), SUGGESTS CONFORMATIONAL CHANGE UPON SUBSTRATE BINDING | | Descriptor: | CALCIUM ION, RIBONUCLEASE T1, VANADATE ION | | Authors: | Kostrewa, D, Choe, H.-W, Heinemann, U, Saenger, W. | | Deposit date: | 1989-05-31 | | Release date: | 1989-10-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of guanosine-free ribonuclease T1, complexed with vanadate (V), suggests conformational change upon substrate binding.
Biochemistry, 28, 1989
|
|
1RVA
 
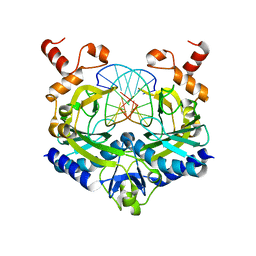 | |
1RVB
 
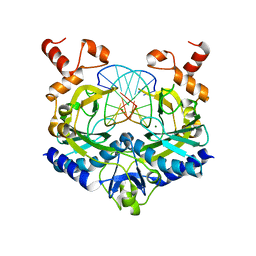 | |
1RVC
 
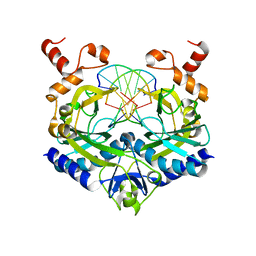 | |
1F97
 
 | | SOLUBLE PART OF THE JUNCTION ADHESION MOLECULE FROM MOUSE | | Descriptor: | JUNCTION ADHESION MOLECULE, MAGNESIUM ION | | Authors: | Kostrewa, D, Brockhaus, M, D'Arcy, A, Dale, G, Bazzoni, G, Dejana, E, Winkler, F, Hennig, M. | | Deposit date: | 2000-07-07 | | Release date: | 2001-01-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of junctional adhesion molecule: structural basis for homophilic adhesion via a novel dimerization motif.
EMBO J., 20, 2001
|
|
1G95
 
 | | CRYSTAL STRUCTURE OF S.PNEUMONIAE GLMU, APO FORM | | Descriptor: | N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE | | Authors: | Kostrewa, D, D'Arcy, A, Kamber, M. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase, GlmU, in apo form at 2.33 A resolution and in complex with UDP-N-acetylglucosamine and Mg(2+) at 1.96 A resolution.
J.Mol.Biol., 305, 2001
|
|
1G97
 
 | | S.PNEUMONIAE GLMU COMPLEXED WITH UDP-N-ACETYLGLUCOSAMINE AND MG2+ | | Descriptor: | MAGNESIUM ION, N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE, SODIUM ION, ... | | Authors: | Kostrewa, D, D'Arcy, A, Kamber, M. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase, GlmU, in apo form at 2.33 A resolution and in complex with UDP-N-acetylglucosamine and Mg(2+) at 1.96 A resolution.
J.Mol.Biol., 305, 2001
|
|
1Z6B
 
 | | Crystal structure of Plasmodium falciparum FabZ at 2.1 A | | Descriptor: | CACODYLATE ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kostrewa, D, Winkler, F.K, Folkers, G, Scapozza, L, Perozzo, R. | | Deposit date: | 2005-03-22 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The crystal structure of PfFabZ, the unique beta-hydroxyacyl-ACP dehydratase involved in fatty acid biosynthesis of Plasmodium falciparum
PROTEIN SCI., 14, 2005
|
|
1AD1
 
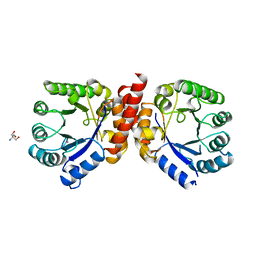 | | DIHYDROPTEROATE SYNTHETASE (APO FORM) FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIHYDROPTEROATE SYNTHETASE, POTASSIUM ION | | Authors: | Kostrewa, D, Oefner, C, D'Arcy, A. | | Deposit date: | 1997-02-19 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the dihydropteroate synthase from Staphylococcus aureus.
J.Mol.Biol., 268, 1997
|
|
4IGF
 
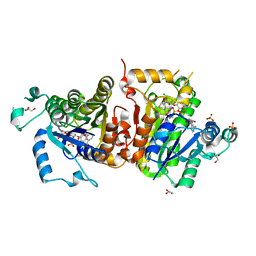 | | Crystal structure of Plasmodium falciparum FabI complexed with NAD and inhibitor 3-(4-Chloro-2-hydroxyphenoxy)-7-hydroxy-2H-chromen-2-one | | Descriptor: | 3-(4-chloro-2-hydroxyphenoxy)-7-hydroxy-2H-chromen-2-one, Enoyl-acyl carrier reductase, GLYCEROL, ... | | Authors: | Kostrewa, D, Perozzo, R. | | Deposit date: | 2012-12-17 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, Synthesis, and Biological and Crystallographic Evaluation of Novel Inhibitors of Plasmodium falciparum Enoyl-ACP-reductase (PfFabI)
J.Med.Chem., 56, 2013
|
|
4IGE
 
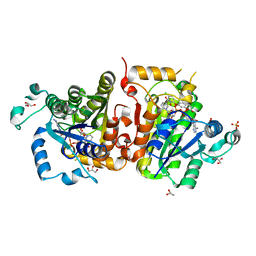 | | Crystal structure of Plasmodium falciparum FabI complexed with NAD and inhibitor 7-(4-Chloro-2-hydroxyphenoxy)-4-methyl-2H-chromen-2-one | | Descriptor: | 7-(4-chloro-2-hydroxyphenoxy)-4-methyl-2H-chromen-2-one, Enoyl-acyl carrier reductase, GLYCEROL, ... | | Authors: | Kostrewa, D, Perozzo, R. | | Deposit date: | 2012-12-17 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design, Synthesis, and Biological and Crystallographic Evaluation of Novel Inhibitors of Plasmodium falciparum Enoyl-ACP-reductase (PfFabI)
J.Med.Chem., 56, 2013
|
|
5N6I
 
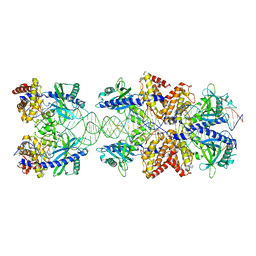 | | Crystal structure of mouse cGAS in complex with 39 bp DNA | | Descriptor: | Cyclic GMP-AMP synthase, DNA (36-MER), DNA (37-MER), ... | | Authors: | Andreeva, L, Kostrewa, D, Hopfner, K.-P. | | Deposit date: | 2017-02-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders.
Nature, 549, 2017
|
|
6FML
 
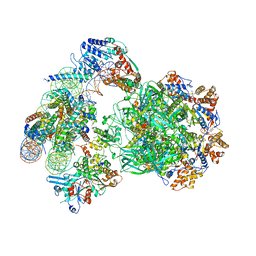 | | CryoEM Structure INO80core Nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin related protein 5, ... | | Authors: | Eustermann, S, Schall, K, Kostrewa, D, Strauss, M, Hopfner, K. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Structural basis for ATP-dependent chromatin remodelling by the INO80 complex.
Nature, 556, 2018
|
|
2GNN
 
 | | Crystal Structure of the Orf Virus NZ2 Variant of VEGF-E | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, ... | | Authors: | Prota, A.E, Pieren, M, Wagner, A, Kostrewa, D, Winkler, F.K, Ballmer-Hofer, K. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-09 | | Last modified: | 2025-11-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Orf virus NZ2 variant of vascular endothelial growth factor-E. Implications for receptor specificity.
J.Biol.Chem., 281, 2006
|
|
2AKF
 
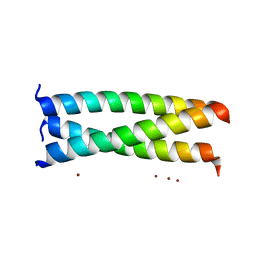 | | Crystal structure of the coiled-coil domain of coronin 1 | | Descriptor: | Coronin-1A, ZINC ION | | Authors: | Kammerer, R.A, Kostrewa, D, Progias, P, Honnappa, S, Avila, D, Lustig, A, Winkler, F.K, Pieters, J, Steinmetz, M.O. | | Deposit date: | 2005-08-03 | | Release date: | 2005-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A conserved trimerization motif controls the topology of short coiled coils
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2XTB
 
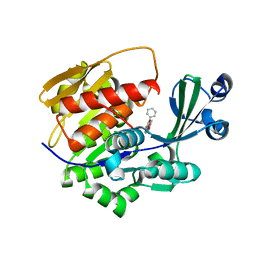 | | Crystal Structure of Trypanosoma brucei rhodesiense Adenosine Kinase Complexed with Activator | | Descriptor: | 4-[5-(4-PHENOXYPHENYL)-1H-PYRAZOL-3-YL]MORPHOLINE, ADENOSINE KINASE | | Authors: | Kuettel, S, Greenwald, J, Kostrewa, D, Ahmed, S, Scapozza, L, Perozzo, R. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of T. B. Rhodesiense Adenosine Kinase Complexed with Inhibitor and Activator: Implications for Catalysis and Hyperactivation.
Plos Negl Trop Dis, 5, 2011
|
|
6FHS
 
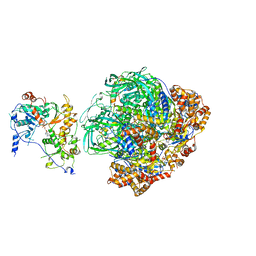 | | CryoEM Structure of INO80core | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Arp5, ... | | Authors: | Eustermann, S, Schall, K, Kostrewa, D, Strauss, M, Hopfner, K. | | Deposit date: | 2018-01-15 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.754 Å) | | Cite: | Structural basis for ATP-dependent chromatin remodelling by the INO80 complex.
Nature, 556, 2018
|
|
2OHF
 
 | |
4C2M
 
 | | Structure of RNA polymerase I at 2.8 A resolution | | Descriptor: | DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA14, ... | | Authors: | Engel, C, Sainsbury, S, Cheung, A.C, Kostrewa, D, Cramer, P. | | Deposit date: | 2013-08-19 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RNA Polymerase I Structure and Transcription Regulation.
Nature, 502, 2013
|
|
5HCN
 
 | | GPN-loop GTPase Npa3 in complex with GMPPCP | | Descriptor: | GLYCEROL, GPN-loop GTPase 1, LAURIC ACID, ... | | Authors: | Niesser, J, Wagner, F.R, Kostrewa, D, Muehlbacher, W, Cramer, P. | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of GPN-Loop GTPase Npa3 and Implications for RNA Polymerase II Assembly.
Mol.Cell.Biol., 36, 2015
|
|
