1G1Y
 
 | | CRYSTAL STRUCTURE OF ALPHA-AMYLASE II (TVAII) FROM THERMOACTINOMYCES VULGARIS R-47 AND BETA-CYCLODEXTRIN COMPLEX | | Descriptor: | ALPHA-AMYLASE II, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Kondo, S, Ohtaki, A, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2000-10-16 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Studies on the hydrolyzing mechanism for cyclodextrins of Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structure of the mutant E354A complexed with beta-cyclodextrin, and kinetic analyses on cyclodextrins.
J.Biochem.(Tokyo), 129, 2001
|
|
1ULZ
 
 | | Crystal structure of the biotin carboxylase subunit of pyruvate carboxylase | | Descriptor: | pyruvate carboxylase n-terminal domain | | Authors: | Kondo, S, Nakajima, Y, Sugio, S, Yong-Biao, J, Sueda, S, Kondo, H. | | Deposit date: | 2003-09-18 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the biotin carboxylase subunit of pyruvate carboxylase from Aquifex aeolicus at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2DZD
 
 | | Crystal structure of the biotin carboxylase domain of pyruvate carboxylase | | Descriptor: | pyruvate carboxylase | | Authors: | Kondo, S, Nakajima, Y, Sugio, S, Sueda, S, Islam, M.N, Kondo, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the biotin carboxylase domain of pyruvate carboxylase from Bacillus thermodenitrificans
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
4W94
 
 | | Crystal structure of cross-linked tetragonal hen egg white lysozyme soaked with 5mM [Ru(CO)3Cl2]2 | | Descriptor: | CHLORIDE ION, DIMETHYLFORMAMIDE, Lysozyme C, ... | | Authors: | Tabe, H, Fujita, K, Abe, S, Tsujimoto, M, Kuchimaru, T, Kizaka-Kondo, S, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Preparation of a Cross-Linked Porous Protein Crystal Containing Ru Carbonyl Complexes as a CO-Releasing Extracellular Scaffold
Inorg.Chem., 54, 2015
|
|
1N5X
 
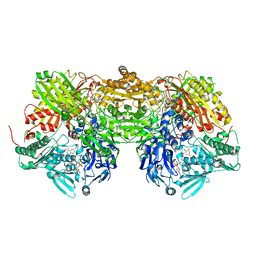 | | Xanthine Dehydrogenase from Bovine Milk with Inhibitor TEI-6720 Bound | | Descriptor: | 2-(3-CYANO-4-ISOBUTOXY-PHENYL)-4-METHYL-5-THIAZOLE-CARBOXYLIC ACID, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Okamoto, K, Eger, B.T, Nishino, T, Kondo, S, Pai, E.F, Nishino, T. | | Deposit date: | 2002-11-07 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An Extremely Potent Inhibitor of Xanthine Oxidoreductase: Crystal Structure of the Enzyme-Inhibitor Complex and Mechanism of Inhibition
J.BIOL.CHEM., 278, 2003
|
|
1BVZ
 
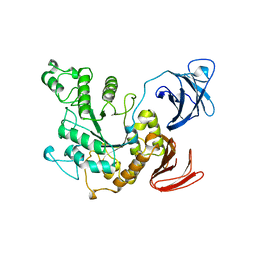 | | ALPHA-AMYLASE II (TVAII) FROM THERMOACTINOMYCES VULGARIS R-47 | | Descriptor: | PROTEIN (ALPHA-AMYLASE II) | | Authors: | Kamitori, S, Kondo, S, Okuyama, K, Yokota, T, Shimura, Y, Tonozuka, T, Sakano, Y. | | Deposit date: | 1998-09-22 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase II (TVAII) hydrolyzing cyclodextrins and pullulan at 2.6 A resolution.
J.Mol.Biol., 287, 1999
|
|
4W96
 
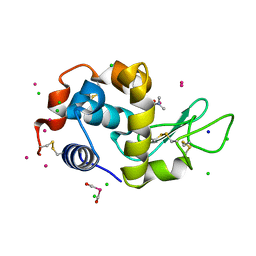 | | Crystal structure of cross-linked tetragonal hen egg white lysozyme soaked with 5mM [Ru(CO)3Cl2]2 followed by the reaction in deoxy-myoglobin solution | | Descriptor: | CHLORIDE ION, DIMETHYLFORMAMIDE, Lysozyme C, ... | | Authors: | Tabe, H, Fujita, K, Abe, S, Tsujimoto, M, Kuchimaru, T, Kizaka-Kondo, S, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Preparation of a Cross-Linked Porous Protein Crystal Containing Ru Carbonyl Complexes as a CO-Releasing Extracellular Scaffold
Inorg.Chem., 54, 2015
|
|
1JF5
 
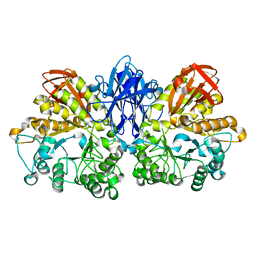 | | CRYSTAL STRUCTURE OF THERMOACTINOMYCES VULGARIS R-47 ALPHA-AMYLASE 2 MUTANT F286A | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
1JF6
 
 | | Crystal structure of thermoactinomyces vulgaris r-47 alpha-amylase mutant F286Y | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
8Z5L
 
 | | Crystal structure of metallo-beta-lactamse, IMP-1, complexed with a quinolinone-based inhibitor | | Descriptor: | 3-[2-azanyl-5-[2-cyclohexylethyl-[3-(4-methylphenoxy)propyl]amino]phenyl]propanoic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Kamo, T, Kuroda, K, Nimura, S, Guo, Y, Kondo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Development of Inhibitory Compounds for Metallo-beta-lactamase through Computational Design and Crystallographic Analysis.
Biochemistry, 63, 2024
|
|
6JKA
 
 | | Crystal structure of metallo-beta-lactamse, IMP-1, in complex with a thiazole-bearing inhibitor | | Descriptor: | 3-[2-azanyl-5-[2-(phenoxymethyl)-1,3-thiazol-4-yl]phenyl]propanoic acid, 6-[2-(phenoxymethyl)-1,3-thiazol-4-yl]-3,4-dihydro-1H-quinolin-2-one, Metallo-beta-lactamase type 2, ... | | Authors: | Kamo, T, Kuroda, K, Kondo, S, Hayashi, U, Fudo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Identification of the Inhibitory Compounds for Metallo-beta-lactamases and Structural Analysis of the Binding Modes.
Chem Pharm Bull (Tokyo), 69, 2021
|
|
6JKB
 
 | | Crystal structure of metallo-beta-lactamse, NDM-1, in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Kamo, T, Kuroda, K, Kondo, S, Hayashi, U, Fudo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Identification of the Inhibitory Compounds for Metallo-beta-lactamases and Structural Analysis of the Binding Modes.
Chem Pharm Bull (Tokyo), 69, 2021
|
|
3AWK
 
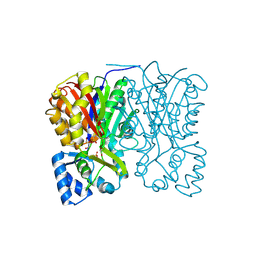 | | Crystal structure of the polyketide synthase 1 from huperzia serrata | | Descriptor: | Chalcone synthase-like polyketide synthase, GLYCEROL, SULFATE ION | | Authors: | Morita, H, Kondo, S, Kato, R, Sugio, S, Kohno, T, Abe, I. | | Deposit date: | 2011-03-23 | | Release date: | 2011-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis of unnatural alkaloid scaffolds by exploiting plant polyketide synthase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AWJ
 
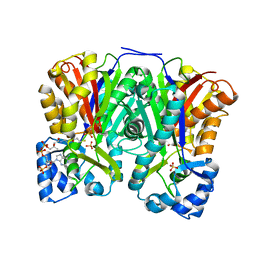 | | Crystal structure of the Huperzia serrata polyketide synthase 1 complexed with CoA-SH | | Descriptor: | COENZYME A, Chalcone synthase-like polyketide synthase, SULFATE ION | | Authors: | Morita, H, Kondo, S, Kato, R, Sugio, S, Kohno, T, Abe, I. | | Deposit date: | 2011-03-23 | | Release date: | 2011-08-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis of unnatural alkaloid scaffolds by exploiting plant polyketide synthase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2D3M
 
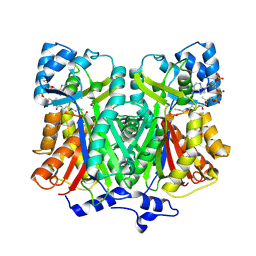 | | Pentaketide chromone synthase complexed with coenzyme A | | Descriptor: | COENZYME A, pentaketide chromone synthase | | Authors: | Morita, H, Kondo, S, Oguro, S, Noguchi, H, Sugio, S, Abe, I, Kohno, T. | | Deposit date: | 2005-09-29 | | Release date: | 2006-10-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insight into Chain-Length Control and Product Specificity of Pentaketide Chromone Synthase from Aloe arborescens
Chem.Biol., 14, 2007
|
|
2D39
 
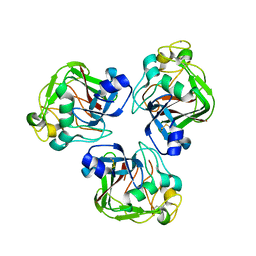 | | Trivalent Recognition Unit of Innate Immunity System; Crystal Structure of human M-ficolin Fibrinogen-like Domain | | Descriptor: | CALCIUM ION, Ficolin-1 | | Authors: | Tanio, M, Kondo, S, Sugio, S, Kohno, T. | | Deposit date: | 2005-09-26 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trivalent recognition unit of innate immunity system; crystal structure of trimeric human m-ficolin fibrinogen-like domain
J.Biol.Chem., 282, 2007
|
|
3WVU
 
 | | Crystal Structure of RuCO/apo-R52CFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Fujita, K, Tanaka, Y, Abe, S, Hikage, T, Kuchimaru, T, Kizaka-Kondoh, S, Ueno, T. | | Deposit date: | 2014-06-09 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Intracellular CO release from composite of ferritin and ruthenium carbonyl complexes.
J.Am.Chem.Soc., 136, 2014
|
|
3WVW
 
 | | Crystal structure of RuCO/apo-WTFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Fujita, K, Tanaka, Y, Abe, S, Hikage, T, Kuchimaru, T, Kizaka-Kondoh, S, Ueno, T. | | Deposit date: | 2014-06-09 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intracellular CO release from composite of ferritin and ruthenium carbonyl complexes.
J.Am.Chem.Soc., 136, 2014
|
|
3WVV
 
 | | Crystal Structure of RuCO/apo-E45C_C48AFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Fujita, K, Tanaka, Y, Abe, S, Hikage, T, Kuchimaru, T, Kizaka-Kondoh, S, Ueno, T. | | Deposit date: | 2014-06-09 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Intracellular CO release from composite of ferritin and ruthenium carbonyl complexes.
J.Am.Chem.Soc., 136, 2014
|
|
6KD5
 
 | | Crystal structure of the extracellular domain of MSPL/TMPRSS13 in complex with dec-RVKR-cmk inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DECANOYL-ARG-VAL-LYS-ARG-CHLOROMETHYLKETONE INHIBITOR, ... | | Authors: | Ohno, A, Maita, N, Okumura, Y, Nikawa, T. | | Deposit date: | 2019-06-30 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of inhibitor-bound human MSPL that can activate high pathogenic avian influenza.
Life Sci Alliance, 4, 2021
|
|
8PBV
 
 | |
5AXI
 
 | | Crystal structure of Cbl-b TKB domain in complex with Cblin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cblin, ... | | Authors: | Ohno, A, Maita, N, Ochi, A, Nakao, R, Nikawa, T. | | Deposit date: | 2015-07-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the TKB domain of ubiquitin ligase Cbl-b complexed with its small inhibitory peptide, Cblin
Arch.Biochem.Biophys., 594, 2016
|
|
7LS3
 
 | | Co-complex CYP46A1 with 8114 (3f) | | Descriptor: | (5-methyl-2-pyridin-4-yl-phenyl)-[4-oxidanyl-4-(phenylmethyl)piperidin-1-yl]methanone, Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lane, W, Yano, J. | | Deposit date: | 2021-02-17 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Soticlestat, a Potent and Selective Inhibitor for Cholesterol 24-Hydroxylase (CH24H).
J.Med.Chem., 64, 2021
|
|
7LRL
 
 | | Co-complex CYP46A1 with 7742 (Soticlestat/TAK-935)) | | Descriptor: | Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, [4-oxidanyl-4-(phenylmethyl)piperidin-1-yl]-(2-pyridin-4-ylpyridin-3-yl)methanone | | Authors: | Lane, W, Yano, J. | | Deposit date: | 2021-02-16 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Discovery of Soticlestat, a Potent and Selective Inhibitor for Cholesterol 24-Hydroxylase (CH24H).
J.Med.Chem., 64, 2021
|
|
7LS4
 
 | | Co-complex CYP46A1 with 9129 (1b) | | Descriptor: | Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, [5,5-dimethyl-3-(2-methylphenyl)-4~{H}-pyrazol-1-yl]-pyridin-4-yl-methanone | | Authors: | Lane, W, Yano, J. | | Deposit date: | 2021-02-17 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Soticlestat, a Potent and Selective Inhibitor for Cholesterol 24-Hydroxylase (CH24H).
J.Med.Chem., 64, 2021
|
|
