7BQC
 
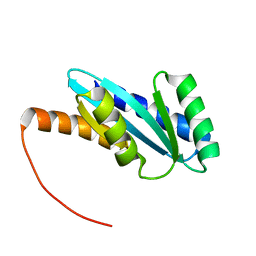 | | Solution NMR structure of NF4; de novo designed protein with a novel fold | | Descriptor: | NF4 | | Authors: | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, T, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2023-07-05 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
2MQ8
 
 | | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414 | | Descriptor: | De novo designed protein LFR1 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-06-12 | | Release date: | 2014-08-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Control over overall shape and size in de novo designed proteins.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2LCI
 
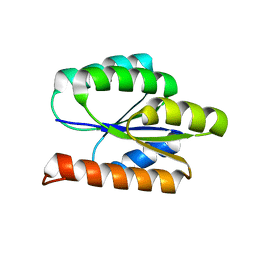 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, P-LOOP NTPASE FOLD, Northeast Structural Genomics Consortium Target OR36 (CASD Target) | | Descriptor: | Protein OR36 | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Janjua, H, Ciccosanti, C, Lee, H, Acton, T.B, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-29 | | Release date: | 2011-06-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target OR36
To be Published
|
|
2LR0
 
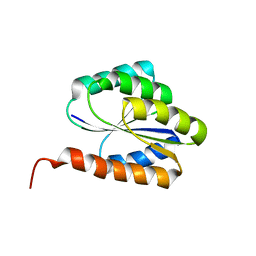 | | Solution NMR structure of de novo designed protein, p-loop ntpase fold, northeast structural genomics consortium target or136 | | Descriptor: | P-loop ntpase fold | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-03-19 | | Release date: | 2012-07-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of de novo designed protein, p-loop ntpase fold, northeast structural genomics consortium target or136
To be Published
|
|
2N3Z
 
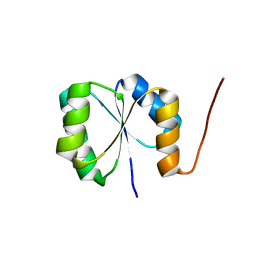 | | Solution NMR Structure of de novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446 | | Descriptor: | OR446 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-15 | | Release date: | 2015-09-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446
To be Published
|
|
2N75
 
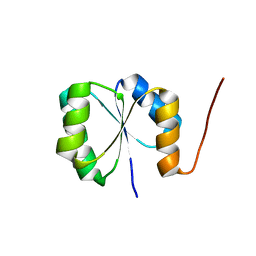 | | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446 | | Descriptor: | De novo designed protein | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446
To be Published
|
|
2N76
 
 | | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414 | | Descriptor: | De novo designed protein LFR1 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414
To be Published
|
|
2MTL
 
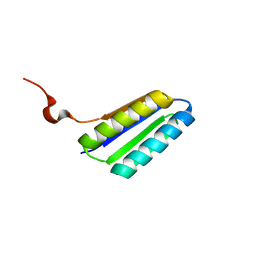 | | Solution NMR Structure of De novo designed FR55, Northeast Structural Genomics Consortium (NESG) Target OR109 | | Descriptor: | De novo designed protein FR55 OR109 | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Ciccosanti, C, Sahdev, S, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed FR55, Northeast Structural Genomics Consortium (NESG) Target OR109
To be Published
|
|
2L82
 
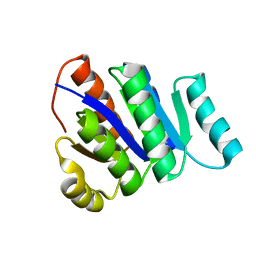 | | Solution NMR Structure of de novo designed protein, P-loop NTPase fold, Northeast Structural Genomics Consortium Target OR32 | | Descriptor: | DESIGNED PROTEIN OR32 | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Janjua, H, Tong, S, Acton, T.B, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-31 | | Release date: | 2011-02-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target OR32
To be Published
|
|
2L69
 
 | | Solution NMR Structure of de novo designed protein, P-loop NTPase fold, Northeast Structural Genomics Consortium Target OR28 | | Descriptor: | Rossmann 2x3 fold protein | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Mao, A, Mao, B, Patel, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-17 | | Release date: | 2011-01-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of de novo designed rossmann 2x3 fold protein, Northeast Structural Genomics Consortium Target OR28
To be Published
|
|
2LND
 
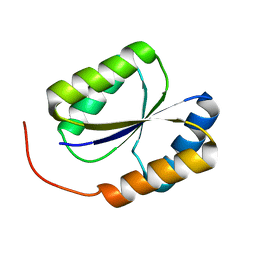 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, PFK fold, Northeast Structural Genomics Consortium Target OR134 | | Descriptor: | DE NOVO DESIGNED PROTEIN, PFK fold | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-23 | | Release date: | 2012-01-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target OR134
To be Published
|
|
2LRH
 
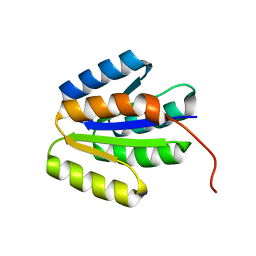 | | Solution NMR structure of de novo designed protein, p-loop ntpase fold, Northeast Structural Genomics Consortium target or137 | | Descriptor: | De novo designed protein | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-03-30 | | Release date: | 2012-07-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of de novo designed protein, p-loop ntpase fold, Northeast Structural Genomics Consortium target or137
To be Published
|
|
2N2T
 
 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN (FDA_60), Northeast Structural Genomics Consortium (NESG) Target OR303 | | Descriptor: | OR303 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Hamilton, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-05-14 | | Release date: | 2015-09-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN (FDA_60), Northeast Structural Genomics Consortium (NESG) Target OR303
To be Published
|
|
2N2U
 
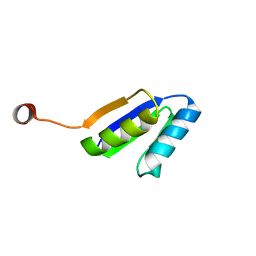 | | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358 | | Descriptor: | OR358 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Hamilton, K, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-05-14 | | Release date: | 2015-09-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358
To be Published
|
|
8IGU
 
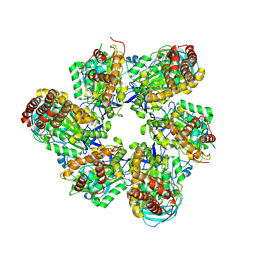 | |
8IGW
 
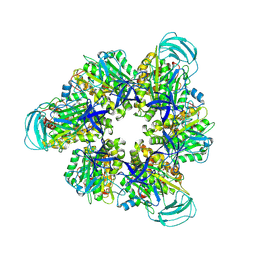 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 4 ADPs: A3(De)3_(ADP)3cat,1non-cat, Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP)3cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
8IGV
 
 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP-Pi)1cat(ADP)2cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
7COQ
 
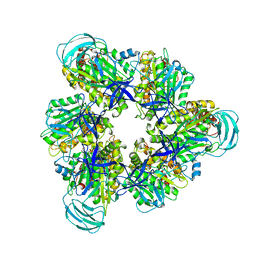 | |
6XEH
 
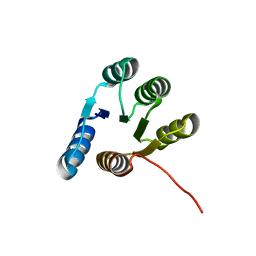 | |
7KBQ
 
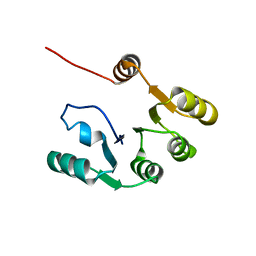 | |
5GAJ
 
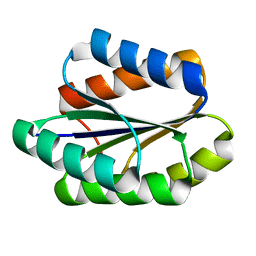 | | Solution NMR structure of De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258 | | Descriptor: | DE NOVO DESIGNED PROTEIN OR258 | | Authors: | Liu, G, Castelllanos, J, Koga, R, Koga, N, Xiao, R, Pederson, K, Janjua, H, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258
To Be Published
|
|
1GD6
 
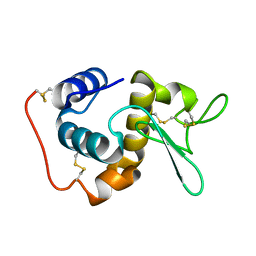 | | STRUCTURE OF THE BOMBYX MORI LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Matsuura, A, Aizawa, T, Yao, M, Kawano, K, Tanaka, I, Nitta, K. | | Deposit date: | 2000-09-19 | | Release date: | 2001-03-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of an insect lysozyme exhibiting catalytic efficiency at low temperatures.
Biochemistry, 41, 2002
|
|
6YW6
 
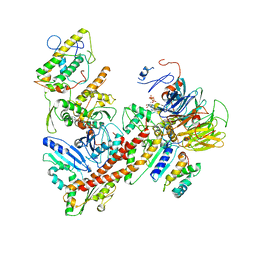 | | Cryo-EM structure of the ARP2/3 1B5CL isoform complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ARPC1B, Actin-related protein 2, ... | | Authors: | von Loeffelholz, O, Moores, C, Purkiss, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of human Arp2/3 complexes provides structural insights into actin nucleation modulation by ARPC5 isoforms.
Biol Open, 9, 2020
|
|
6YW7
 
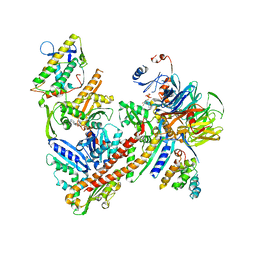 | | Cryo-EM structure of the ARP2/3 1A5C isoform complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1A, ... | | Authors: | von Loeffelholz, O, Moores, C, Purkiss, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM of human Arp2/3 complexes provides structural insights into actin nucleation modulation by ARPC5 isoforms.
Biol Open, 9, 2020
|
|
