3SM0
 
 | | tRNA-Guanine Transglycosylase in complex with lin-Benzohypoxanthine Inhibitor | | Descriptor: | 4-{2-[(cyclopentylmethyl)amino]ethyl}-2-(methylamino)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Klebe, G, Immekus, F, Heine, A. | | Deposit date: | 2011-06-27 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | From lin-Benzoguanines to lin-Benzohypoxanthines as Ligands for Zymomonas mobilis tRNA-Guanine Transglycosylase: Replacement of Protein-Ligand Hydrogen Bonding by Importing Water Clusters.
Chemistry, 18, 2012
|
|
3TLL
 
 | | tRNA-Guanine Transglycosylase in complex with N-Ethyl-lin-benzoguanine Inhibitor | | Descriptor: | 6-(ethylamino)-2-(methylamino)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Klebe, G, Immekus, F, Heine, A. | | Deposit date: | 2011-08-30 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | From lin-Benzoguanines to lin-Benzohypoxanthines as Ligands for Zymomonas mobilis tRNA-Guanine Transglycosylase: Replacement of Protein-Ligand Hydrogen Bonding by Importing Water Clusters.
Chemistry, 18, 2012
|
|
3S1G
 
 | | tRNA-Guanine Transglycosylase in complex with lin-Benzohypoxanthine Inhibitor | | Descriptor: | 2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Klebe, G, Immekus, F, Heine, A. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | From lin-Benzoguanines to lin-Benzohypoxanthines as Ligands for Zymomonas mobilis tRNA-Guanine Transglycosylase: Replacement of Protein-Ligand Hydrogen Bonding by Importing Water Clusters.
Chemistry, 18, 2012
|
|
2Q1B
 
 | | Carbonic Anhydrase II in Complex with Saccharin | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Klebe, G, Heine, A, Koehler, K. | | Deposit date: | 2007-05-24 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Saccharin Inhibits Carbonic Anhydrases: Possible Explanation for its Unpleasant Metallic Aftertaste.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
3RR4
 
 | | tRNA-Guanine Transglycosylase in complex with N-Methyl-lin-Benzoguanine Inhibitor | | Descriptor: | 2,6-bis(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Klebe, G, Immekus, F, Heine, A. | | Deposit date: | 2011-04-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | From lin-Benzoguanines to lin-Benzohypoxanthines as Ligands for Zymomonas mobilis tRNA-Guanine Transglycosylase: Replacement of Protein-Ligand Hydrogen Bonding by Importing Water Clusters.
Chemistry, 18, 2012
|
|
2AGT
 
 | | Aldose Reductase Mutant Leu 300 Pro complexed with Fidarestat | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | Authors: | Petrova, T, Steuber, H, Hazemann, I, Cousido-Siah, A, Mitschler, A, Chung, R, Oka, M, Klebe, G, El-Kabbani, O, Joachimiak, A, Podjarny, A. | | Deposit date: | 2005-07-27 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Factorizing Selectivity Determinants of Inhibitor Binding toward Aldose and Aldehyde Reductases: Structural and Thermodynamic Properties of the Aldose Reductase Mutant Leu300Pro-Fidarestat Complex
J.Med.Chem., 48, 2005
|
|
3KIG
 
 | | Mutant carbonic anhydrase II in complex with an azide and an alkyne | | Descriptor: | 2-azido-N-(2-sulfanylethyl)ethanamide, 3-ethynylbenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Schulze-Wischeler, J, Niehage, N.U, Heine, A, Klebe, G. | | Deposit date: | 2009-11-02 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Stereo- and Regioselective Azide/Alkyne Cycloadditions in Carbonic Anhydrase II via Tethering, Monitored by Crystallography and Mass Spectrometry.
Chemistry, 17, 2011
|
|
3KNE
 
 | | Carbonic Anhydrase II H64C mutant in complex with an in situ formed triazole | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, Carbonic anhydrase 2, N-[(S)-(1-{2-oxo-2-[(3-sulfanylpropyl)amino]ethyl}-1H-1,2,3-triazol-5-yl)(phenyl)methyl]-4-sulfamoylbenzamide, ... | | Authors: | Schulze-Wischeler, J, Heine, A, Klebe, G. | | Deposit date: | 2009-11-12 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Stereo- and Regioselective Azide/Alkyne Cycloadditions in Carbonic Anhydrase II via Tethering, Monitored by Crystallography and Mass Spectrometry.
Chemistry, 17, 2011
|
|
4LAP
 
 | |
4LEQ
 
 | | tRNA guanine transglycosylase (TGT) in complex with Furanoside-Based lin-Benzoguanine 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2013-06-26 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.405 Å) | | Cite: | Replacement of Water Molecules in a Phosphate Binding Site by Furanoside-Appended lin-Benzoguanine Ligands of tRNA-Guanine Transglycosylase (TGT).
Chemistry, 21, 2015
|
|
4D91
 
 | |
4D9W
 
 | |
7APM
 
 | | tRNA-guanine transglycosylase H319C mutant spin-labeled with MTSL. | | Descriptor: | CHLORIDE ION, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Nguyen, D, Heine, A, Klebe, G. | | Deposit date: | 2020-10-18 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unraveling a Ligand-Induced Twist of a Homodimeric Enzyme by Pulsed Electron-Electron Double Resonance.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7APL
 
 | |
6ERT
 
 | |
6F14
 
 | | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with isoquinoline and PKI (5-24) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ISOQUINOLINE, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2017-11-21 | | Release date: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (1.867 Å) | | Cite: | Fragment based drug design - Small chemical changes of fragments effecting big changes in binding
To Be Published
|
|
3RML
 
 | | Human Thrombin in complex with MI331 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
3RLY
 
 | | Human Thrombin in complex with MI329 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
3RMN
 
 | | Human Thrombin in complex with MI341 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
3S3J
 
 | |
8CNP
 
 | |
8CNF
 
 | |
2BBF
 
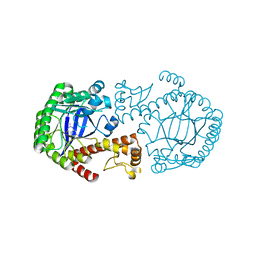 | | Crystal structure of tRNA-guanine transglycosylase (TGT) from Zymomonas mobilis in complex with 6-amino-3,7-dihydro-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-3,7-DIHYDRO-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, ZINC ION, tRNA guanine transglycosylase | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2005-10-17 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding.
J.Mol.Biol., 370, 2007
|
|
7NHW
 
 | |
7NL8
 
 | |
