1I07
 
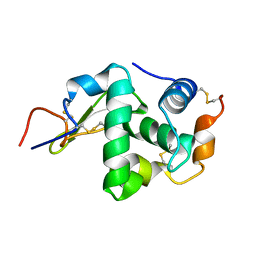
 | | EPS8 SH3 DOMAIN INTERTWINED DIMER | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR KINASE SUBSTRATE EPS8 | | Authors: | Kishan, K.V.R, Newcomer, M.E. | | Deposit date: | 2001-01-29 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of pH and salt bridges on structural assembly: molecular structures of the monomer and intertwined dimer of the Eps8 SH3 domain.
Protein Sci., 10, 2001
|
|
1I0C
 
 | | EPS8 SH3 CLOSED MONOMER | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR KINASE SUBSTRATE EPS8 | | Authors: | Kishan, K.V.R, Newcomer, M.E. | | Deposit date: | 2001-01-29 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effect of pH and salt bridges on structural assembly: molecular structures of the monomer and intertwined dimer of the Eps8 SH3 domain.
Protein Sci., 10, 2001
|
|
3ZVQ
 
 | | Crystal Structure of proteolyzed lysozyme | | Descriptor: | LYSOZYME C | | Authors: | Kishan, K.V, Sharma, A. | | Deposit date: | 2011-07-26 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Salt-assisted religation of proteolyzed Glutathione-S-transferase follows Hofmeister series.
Protein Pept.Lett., 17, 2010
|
|
1AOJ
 
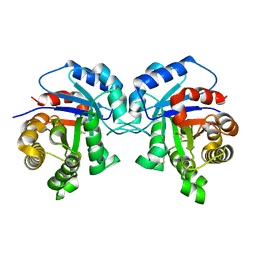
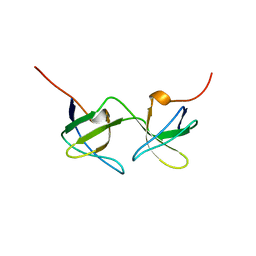 | |
1TPF
 
 | |
1TPE
 
 | |
1MSS
 
 | |
1YNY
 
 | | Molecular Structure of D-Hydantoinase from a Bacillus sp. AR9: Evidence for mercury inhibition | | Descriptor: | D-Hydantoinase, MANGANESE (II) ION | | Authors: | Radha Kishan, K.V, Vohra, R.M, Ganeshan, K, Agrawal, V, Sharma, V.M, Sharma, R. | | Deposit date: | 2005-01-26 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular structure of D-hydantoinase from Bacillus sp. AR9: evidence for mercury inhibition.
J.Mol.Biol., 347, 2005
|
|
1TTJ
 
 | |
1TTI
 
 | |
4KQN
 
 | |
4KIR
 
 | |
1ML1
 
 | | PROTEIN ENGINEERING WITH MONOMERIC TRIOSEPHOSPHATE ISOMERASE: THE MODELLING AND STRUCTURE VERIFICATION OF A SEVEN RESIDUE LOOP | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Thanki, N, Zeelen, J.P, Mathieu, M, Jaenicke, R, Abagyan, R.A, Wierenga, R, Schliebs, W. | | Deposit date: | 1996-09-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein engineering with monomeric triosephosphate isomerase (monoTIM): the modelling and structure verification of a seven-residue loop.
Protein Eng., 10, 1997
|
|
1RHB
 
 | | WATER DEPENDENT DOMAIN MOTION AND FLEXIBILITY IN RIBONUCLEASE A AND THE INVARIANT FEATURES IN ITS HYDRATION SHELL. AN X-RAY STUDY OF TWO LOW HUMIDITY CRYSTAL FORMS OF THE ENZYME | | Descriptor: | RIBONUCLEASE A | | Authors: | Radha Kishan, K.V, Chandra, N.R, Sudarsanakumar, C, Suguna, K, Vijayan, M. | | Deposit date: | 1994-11-13 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water-dependent domain motion and flexibility in ribonuclease A and the invariant features in its hydration shell. An X-ray study of two low-humidity crystal forms of the enzyme.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1RHA
 
 | | WATER DEPENDENT DOMAIN MOTION AND FLEXIBILITY IN RIBONUCLEASE A AND THE INVARIANT FEATURES IN ITS HYDRATION SHELL. AN X-RAY STUDY OF TWO LOW HUMIDITY CRYSTAL FORMS OF THE ENZYME | | Descriptor: | RIBONUCLEASE A | | Authors: | Radha Kishan, K.V, Chandra, N.R, Sudarsanakumar, C, Suguna, K, Vijayan, M. | | Deposit date: | 1994-11-13 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Water-dependent domain motion and flexibility in ribonuclease A and the invariant features in its hydration shell. An X-ray study of two low-humidity crystal forms of the enzyme.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
3K1D
 
 | | Crystal structure of glycogen branching enzyme synonym: 1,4-alpha-D-glucan:1,4-alpha-D-GLUCAN 6-glucosyl-transferase from mycobacterium tuberculosis H37RV | | Descriptor: | 1,4-alpha-glucan-branching enzyme | | Authors: | Pal, K, Kumar, S, Swaminathan, K. | | Deposit date: | 2009-09-27 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of full-length Mycobacterium tuberculosis H37Rv glycogen branching enzyme: insights of N-terminal beta-sandwich in substrate specificity and enzymatic activity
J.Biol.Chem., 285, 2010
|
|
1TMH
 
 | |
1TRI
 
 | |
1DKW
 
 | | CRYSTAL STRUCTURE OF TRIOSE-PHOSPHATE ISOMERASE WITH MODIFIED SUBSTRATE BINDING SITE | | Descriptor: | TERTIARY-BUTYL ALCOHOL, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Norledge, B.V, Lambeir, A.M, Abagyan, R.A, Rottman, A, Fernandez, A.M, Filimonov, V.V, Peter, M.G, Wierenga, R.K. | | Deposit date: | 1999-12-08 | | Release date: | 2000-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Modeling, mutagenesis, and structural studies on the fully conserved phosphate-binding loop (loop 8) of triosephosphate isomerase: toward a new substrate specificity.
Proteins, 42, 2001
|
|
1XPT
 
 | | BOVINE RIBONUCLEASE A (PHOSPHATE-FREE) | | Descriptor: | RIBONUCLEASE A | | Authors: | Sadasivan, C, Nagendra, H.G, Vijayan, M. | | Deposit date: | 1998-02-23 | | Release date: | 1998-05-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plasticity, hydration and accessibility in ribonuclease A. The structure of a new crystal form and its low-humidity variant.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1XPS
 
 | |
