1UM8
 
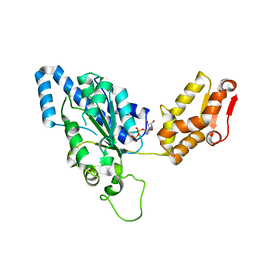 | | Crystal structure of helicobacter pylori ClpX | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit clpX | | Authors: | Kim, D.Y, Kim, K.K. | | Deposit date: | 2003-09-25 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of ClpX Molecular Chaperone from Helicobacter pylori
J.Biol.Chem., 278, 2003
|
|
2L4M
 
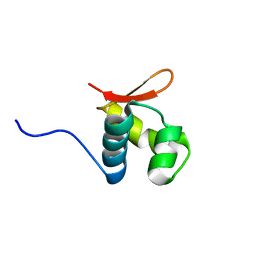 | |
1L1J
 
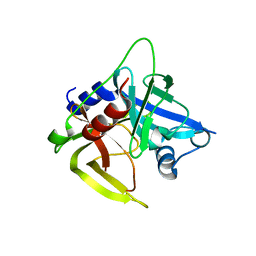 | | Crystal structure of the protease domain of an ATP-independent heat shock protease HtrA | | Descriptor: | heat shock protease HtrA | | Authors: | Kim, D.Y, Kim, D.R, Ha, S.C, Lokanath, N.K, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2002-02-18 | | Release date: | 2003-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Protease Domain of a Heat-shock Protein HtrA from Thermotoga maritima
J.BIOL.CHEM., 278, 2003
|
|
1SFU
 
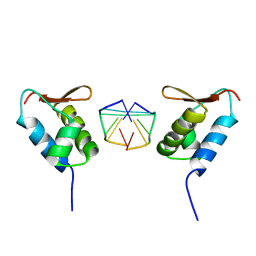 | | Crystal structure of the viral Zalpha domain bound to left-handed Z-DNA | | Descriptor: | 34L protein, 5'-D(*T*CP*GP*CP*GP*CP*G)-3' | | Authors: | Ha, S.C, Van Quyen, D, Wu, C.A, Lowenhaupt, K, Rich, A, Kim, Y.G, Kim, K.K. | | Deposit date: | 2004-02-20 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zalpha bound to DNA.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4NJR
 
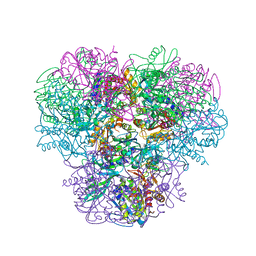 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | CARBONATE ION, Probable M18 family aminopeptidase 2, ZINC ION | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.Y, Ha, S.C, Yun, K.H, Kim, K.S, Kim, J.H, Kim, K.K. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
3M4W
 
 | | Structural basis for the negative regulation of bacterial stress response by RseB | | Descriptor: | Sigma-E factor negative regulatory protein, Sigma-E factor regulatory protein rseB, ZINC ION | | Authors: | Kim, D.Y, Kwon, E, Choi, J.K, Hwang, H.-Y, Kim, K.K. | | Deposit date: | 2010-03-12 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the negative regulation of bacterial stress response by RseB
Protein Sci., 19, 2010
|
|
4P6B
 
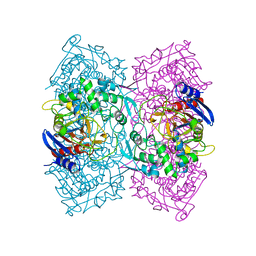 | | Crystal structure of Est-Y29,a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-24 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3OEO
 
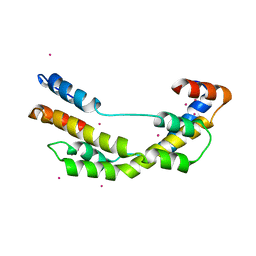 | | The crystal structure E. coli Spy | | Descriptor: | CADMIUM ION, Spheroplast protein Y | | Authors: | Kwon, E, Kim, D.Y, Gross, C.A, Gross, J.D, Kim, K.K. | | Deposit date: | 2010-08-13 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure Escherichia coli Spy.
Protein Sci., 19, 2010
|
|
4P87
 
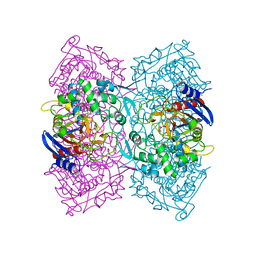 | | Crystal structure of Est-Y29, a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | 4-NITROPHENYL PHOSPHATE, Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-30 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4P85
 
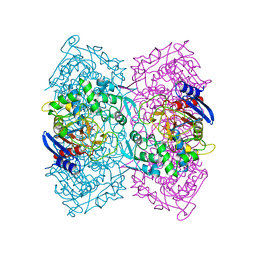 | | Crystal structure of Est-Y29, a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | DIETHYL PHOSPHONATE, Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-30 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3EYI
 
 | |
4YGQ
 
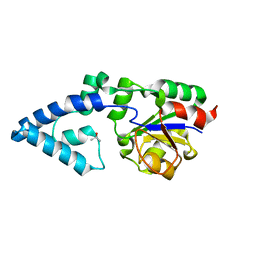 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | Hydrolase, TERTIARY-BUTYL ALCOHOL | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4YGR
 
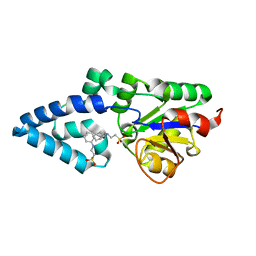 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hydrolase, MAGNESIUM ION | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4YGS
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | CITRIC ACID, Hydrolase, MAGNESIUM ION | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
2P52
 
 | |
2P4B
 
 | | Crystal structure of E.coli RseB | | Descriptor: | Sigma-E factor regulatory protein rseB, octyl beta-D-glucopyranoside | | Authors: | Kim, D.Y, Kim, K.K. | | Deposit date: | 2007-03-12 | | Release date: | 2007-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of RseB and a model of its binding mode to RseA
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4KMF
 
 | |
1PCV
 
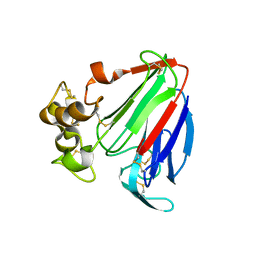 | | Crystal structure of osmotin, a plant antifungal protein | | Descriptor: | osmotin | | Authors: | Min, K, Ha, S.C, Yun, D.-J, Bressan, R.A, Kim, K.K. | | Deposit date: | 2003-05-16 | | Release date: | 2004-02-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of osmotin, a plant antifungal protein
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
3SH5
 
 | |
3SH4
 
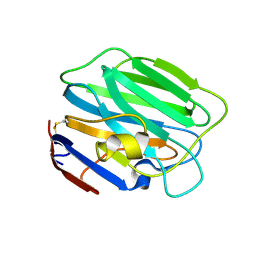 | |
3F21
 
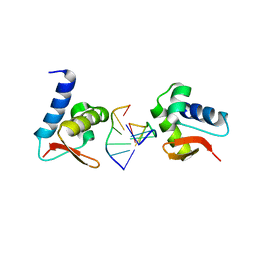 | | Crystal structure of Zalpha in complex with d(CACGTG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DAP*DCP*DGP*DTP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
3F23
 
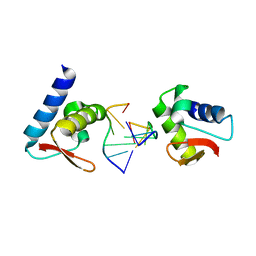 | | Crystal structure of Zalpha in complex with d(CGGCCG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DGP*DGP*DCP*DCP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
3F22
 
 | | Crystal structure of Zalpha in complex with d(CGTACG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DGP*DTP*DAP*DCP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
3U7M
 
 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | Descriptor: | N-((2R,4S)-2-butyl-4-(3-(2-fluorophenyl)ureido)-5-methyl-3-oxohexyl)-N-hydroxyformamide, Peptide deformylase, ZINC ION | | Authors: | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | Deposit date: | 2011-10-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3QBQ
 
 | | Crystal structure of extracellular domains of mouse RANK-RANKL complex | | Descriptor: | Tumor necrosis factor ligand superfamily member 11, Tumor necrosis factor receptor superfamily member 11A | | Authors: | Ta, H.M, Nguyen, G.T.T, Jin, H.M, Choi, J.K, Park, H, Kim, N.S, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2011-01-13 | | Release date: | 2011-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based development of a receptor activator of nuclear factor-kappaB ligand (RANKL) inhibitor peptide and molecular basis for osteopetrosis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
