3QY8
 
 | | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases | | Descriptor: | FE (III) ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, H.S, Lee, S.J, Yoon, H.J, An, D.R, Kim, D.J, Kim, S.-J, Suh, S.W. | | Deposit date: | 2011-03-03 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases.
J.Struct.Biol., 175, 2011
|
|
3QY7
 
 | | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases | | Descriptor: | FE (III) ION, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kim, H.S, Lee, S.J, Yoon, H.J, An, D.R, Kim, D.J, Kim, S.-J, Suh, S.W. | | Deposit date: | 2011-03-03 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases.
J.Struct.Biol., 175, 2011
|
|
3QY6
 
 | | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases | | Descriptor: | FE (III) ION, MAGNESIUM ION, Tyrosine-protein phosphatase YwqE | | Authors: | Kim, H.S, Lee, S.J, Yoon, H.J, An, D.R, Kim, D.J, Kim, S.-J, Suh, S.W. | | Deposit date: | 2011-03-03 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases.
J.Struct.Biol., 175, 2011
|
|
3MYP
 
 | | Crystal structure of tagatose-1,6-bisphosphate aldolase from Staphylococcus aureus | | Descriptor: | Tagatose 1,6-diphosphate aldolase | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Suh, S.W. | | Deposit date: | 2010-05-10 | | Release date: | 2011-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structures of LacD from Staphylococcus aureus and LacD.1 from Streptococcus pyogenes: Insights into substrate specificity and virulence gene regulation
Febs Lett., 585, 2011
|
|
3MYO
 
 | | Crystal structure of tagatose-1,6-bisphosphate aldolase from Streptococcus pyogenes | | Descriptor: | Tagatose 1,6-diphosphate aldolase 1 | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Suh, S.W. | | Deposit date: | 2010-05-10 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of LacD from Staphylococcus aureus and LacD.1 from Streptococcus pyogenes: Insights into substrate specificity and virulence gene regulation
Febs Lett., 585, 2011
|
|
2F5G
 
 | | Crystal structure of IS200 transposase | | Descriptor: | Transposase, putative | | Authors: | Lee, H.H, Yoon, J.Y, Kim, H.S, Kang, J.Y, Kim, K.H, Kim, D.J, Suh, S.W. | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Metal Ion-bound IS200 Transposase
J.Biol.Chem., 281, 2006
|
|
2F4F
 
 | | Crystal structure of IS200 transposase | | Descriptor: | MANGANESE (II) ION, Transposase, putative | | Authors: | Lee, H.H, Yoon, J.Y, Kim, H.S, Kang, J.Y, Kim, K.H, Kim, D.J, Suh, S.W. | | Deposit date: | 2005-11-23 | | Release date: | 2005-12-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Metal Ion-bound IS200 Transposase
J.Biol.Chem., 281, 2006
|
|
2O4C
 
 | | Crystal Structure of D-Erythronate-4-phosphate Dehydrogenase Complexed with NAD | | Descriptor: | Erythronate-4-phosphate dehydrogenase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Ha, J.Y, Lee, J.H, Kim, K.H, Kim, D.J, Lee, H.H, Kim, H.K, Yoon, H.J, Suh, S.W. | | Deposit date: | 2006-12-04 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of d-Erythronate-4-phosphate Dehydrogenase Complexed with NAD
J.Mol.Biol., 366, 2007
|
|
1WOR
 
 | | Crystal Structure of T-protein of the Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, DIHYDROLIPOIC ACID | | Authors: | Lee, H.H, Kim, D.J, Ahn, H.J, Ha, J.Y, Suh, S.W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of T-protein of the Glycine Cleavage System: Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia
J.Biol.Chem., 279, 2004
|
|
1WOP
 
 | | Crystal Structure of T-protein of the Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid | | Authors: | Lee, H.H, Kim, D.J, Ahn, H.J, Ha, J.Y, Suh, S.W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of T-protein of the Glycine Cleavage System: Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia
J.Biol.Chem., 279, 2004
|
|
1WOS
 
 | | Crystal Structure of T-protein of the Glycine Cleavage System | | Descriptor: | Aminomethyltransferase | | Authors: | Lee, H.H, Kim, D.J, Ahn, H.J, Ha, J.Y, Suh, S.W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of T-protein of the Glycine Cleavage System: Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia
J.Biol.Chem., 279, 2004
|
|
1WOO
 
 | | Crystal structure of T-protein of the Glycine Cleavage System | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, Aminomethyltransferase | | Authors: | Lee, H.H, Kim, D.J, Ahn, H.J, Ha, J.Y, Suh, S.W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of T-protein of the Glycine Cleavage System: Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia
J.Biol.Chem., 279, 2004
|
|
3UY5
 
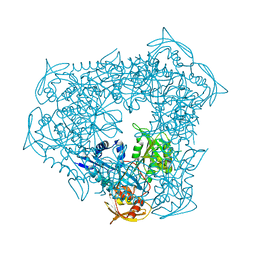 | | crystal structure of Eis from Mycobacterium tuberculosis | | Descriptor: | Enhanced intracellular survival protein | | Authors: | Kim, K.H, Suh, S.W. | | Deposit date: | 2011-12-05 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 2012
|
|
8G0M
 
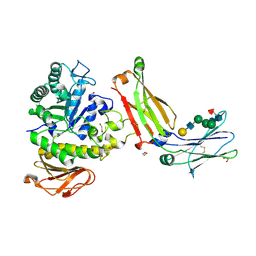 | | Structure of complex between TV6.6 and CD98hc ECD | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, TETRAETHYLENE GLYCOL, ... | | Authors: | Kariolis, M.S, Lexa, K, Liau, N.P.D, Srivastava, D, Tran, H, Wells, R.C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CD98hc is a target for brain delivery of biotherapeutics.
Nat Commun, 14, 2023
|
|
2QI2
 
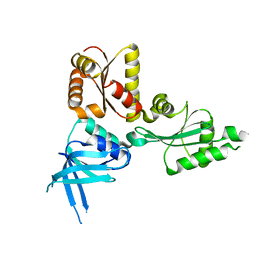 | | Crystal structure of the Thermoplasma acidophilum Pelota protein | | Descriptor: | Cell division protein pelota related protein | | Authors: | Lee, H.H, Kim, Y.S, Kim, K.H, Heo, I.H, Kim, S.K, Kim, O, Suh, S.W. | | Deposit date: | 2007-07-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into dom34, a key component of no-go mRNA decay
Mol.Cell, 27, 2007
|
|
3F3M
 
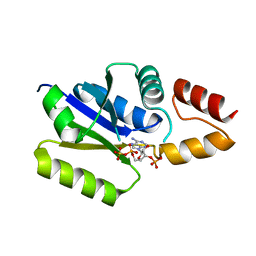 | |
7QJM
 
 | |
7QJN
 
 | | Crystal structure of an alpha/beta-hydrolase enzyme from Candidatus Kryptobacter tengchongensis (306) | | Descriptor: | Dienelactone hydrolase, PHOSPHATE ION | | Authors: | Zahn, M, Gill, R.S, Erickson, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJO
 
 | |
7QJQ
 
 | | Crystal structure of a cutinase enzyme from Thermobifida fusca NTU22 (702) | | Descriptor: | Acetylxylan esterase, DI(HYDROXYETHYL)ETHER | | Authors: | Zahn, M, Gill, R.S, Avilan, L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJR
 
 | | Crystal structure of cutinase 1 from Thermobifida fusca DSM44342 (703) | | Descriptor: | Cutinase 1, TETRAETHYLENE GLYCOL | | Authors: | Zahn, M, Avilan, L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJT
 
 | | Crystal structure of a cutinase enzyme from Thermobifida cellulosilytica TB100 (711) | | Descriptor: | GLYCEROL, MAGNESIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Zahn, M, Shakespeare, T.J, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJP
 
 | | Crystal structure of a cutinase enzyme from Saccharopolyspora flava (611) | | Descriptor: | Cutinase, TETRAETHYLENE GLYCOL | | Authors: | Zahn, M, Avilan, L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJS
 
 | | Crystal structure of a cutinase enzyme from Thermobifida fusca YX (705) | | Descriptor: | Cutinase 2, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zahn, M, Shakespeare, T.J, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
2OWY
 
 | |
