8FZ9
 
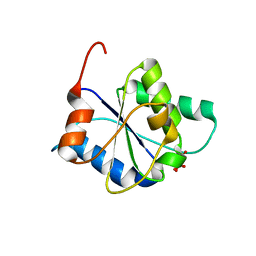 | |
8TCI
 
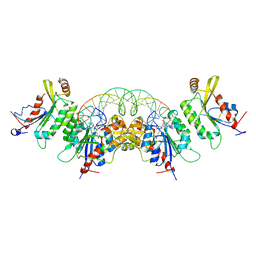 | | Crystal structure of DNMT3C-DNMT3L in complex with CGG DNA | | Descriptor: | (5'-D(P*CP*AP*TP*G)-R(P*(PYO))-D(P*GP*GP*TP*CP*TP*AP*AP*TP*TP*AP*GP*AP*CP*CP*GP*CP*AP*TP*G)-3'), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3C, ... | | Authors: | Khudaverdyan, N, Song, J. | | Deposit date: | 2023-07-01 | | Release date: | 2024-08-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The structure of DNA methyltransferase DNMT3C reveals an activity-tuning mechanism for DNA methylation.
J.Biol.Chem., 300, 2024
|
|
7UBU
 
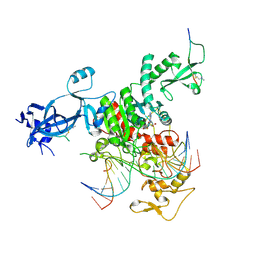 | |
7LMK
 
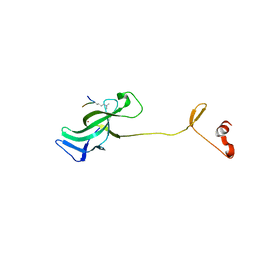 | |
7LMM
 
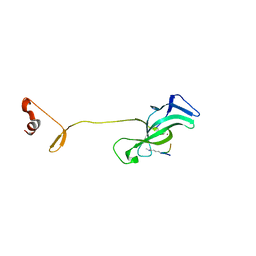 | |
7JLT
 
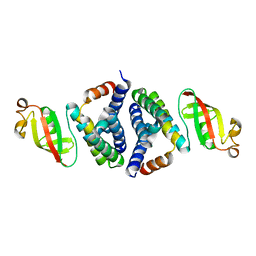 | | Crystal Structure of SARS-CoV-2 NSP7-NSP8 complex. | | Descriptor: | Non-structural protein 7, Non-structural protein 8 | | Authors: | Biswal, M, Hai, R, Song, J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two conserved oligomer interfaces of NSP7 and NSP8 underpin the dynamic assembly of SARS-CoV-2 RdRP.
Nucleic Acids Res., 49, 2021
|
|
9BAP
 
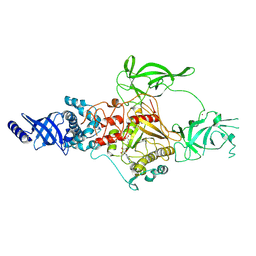 | | CryoEM structure of Apo-DIM2 | | Descriptor: | DNA (cytosine-5-)-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Nat Commun, 15, 2024
|
|
9BAZ
 
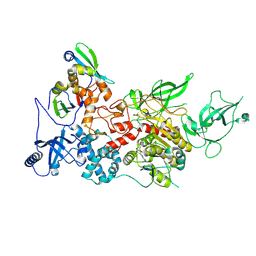 | | CryoEM structure of DIM2-HP1 complex | | Descriptor: | DNA (cytosine-5-)-methyltransferase, Heterochromatin protein one, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-05 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Nat Commun, 15, 2024
|
|
9BAQ
 
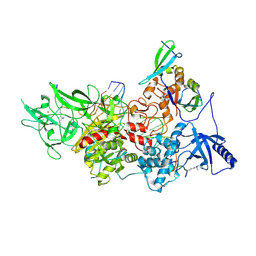 | | CryoEM structure of DIM2-HP1-H3K9me3-DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*CP*T)-R(P*(PYO))-D(P*CP*TP*CP*CP*TP*CP*CP*TP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*TP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*TP*AP*GP*T)-3'), DNA (cytosine-5-)-methyltransferase, ... | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Nat Commun, 15, 2024
|
|
8EIH
 
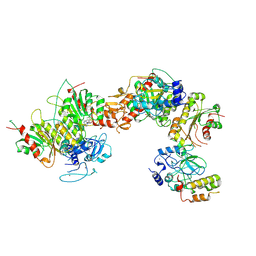 | | Cryo-EM structure of human DNMT3B homo-tetramer (form I) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8EIK
 
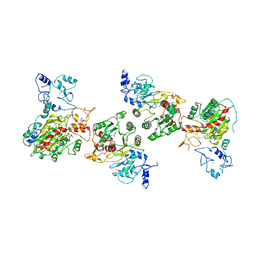 | | Cryo-EM structure of human DNMT3B homo-hexamer | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8EII
 
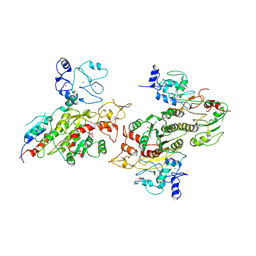 | | Cryo-EM structure of human DNMT3B homo-tetramer (form II) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
8EIJ
 
 | | Cryo-EM structure of human DNMT3B homo-trimer | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Lu, J.W, Song, J.K. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis for the allosteric regulation and dynamic assembly of DNMT3B.
Nucleic Acids Res., 51, 2023
|
|
