2WB6
 
 | | Crystal structure of AFV1-102, a protein from the Acidianus Filamentous Virus 1 | | Descriptor: | AFV1-102, CHLORIDE ION | | Authors: | Keller, J, Leulliot, N, Collinet, B, Campanacci, V, Cambillau, C, Pranghisvilli, D, van Tilbeurgh, H. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Afv1-102, a Protein from the Acidianus Filamentous Virus 1.
Protein Sci., 18, 2009
|
|
2WB7
 
 | | pT26-6p | | Descriptor: | PT26-6P | | Authors: | Keller, J, Leulliot, N, Soler, N, Collinet, B, Vincentelli, R, Forterre, P, van Tilbeurgh, H. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Protein Encoded by a New Family of Mobile Elements from Euryarchaea Exhibits Three Domains with Novel Folds.
Protein Sci., 18, 2009
|
|
2J6C
 
 | | crystal structure of AFV3-109, a highly conserved protein from crenarchaeal viruses | | Descriptor: | AFV3-109, GLYCEROL | | Authors: | Keller, J, Leulliot, N, Cambillau, C, Campanacci, V, Porciero, S, Prangishvili, D, Cortez, D, Quevillon-Cheruel, S, Van Tilbeurgh, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Afv3-109, a Highly Conserved Protein from Crenarchaeal Viruses.
Virol J., 4, 2007
|
|
2J6B
 
 | | crystal structure of AFV3-109, a highly conserved protein from crenarchaeal viruses | | Descriptor: | AFV3-109 | | Authors: | Keller, J, Leulliot, N, Cambillau, C, Campanacci, V, Porciero, S, Prangishvili, D, Cortez, D, Quevillon-Cheruel, S, Van Tilbeurgh, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Afv3-109, a Highly Conserved Protein from Crenarchaeal Viruses.
Virol J., 4, 2007
|
|
4OJJ
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P212121) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-21 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
1S0F
 
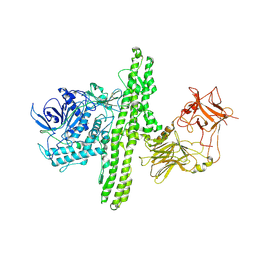 | | Crystal structure of botulinum neurotoxin type B at pH 7.0 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION, ZINC ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
1S0E
 
 | | Crystal structure of botulinum neurotoxin type B at pH 6.0 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION, ZINC ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
4OGP
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
4NFA
 
 | | Structure of the C-terminal doamin of Knl1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein CASC5 | | Authors: | Petrovic, A, Mosalaganti, S, Keller, J, Mattiuzzo, M, Overlack, K, Wohlgemuth, S, Pasqualato, S, Raunser, S, Musacchio, A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Modular Assembly of RWD Domains on the Mis12 Complex Underlies Outer Kinetochore Organization.
Mol.Cell, 53, 2014
|
|
4NF9
 
 | | Structure of the Knl1/Nsl1 complex | | Descriptor: | CHLORIDE ION, Kinetochore-associated protein NSL1 homolog, Protein CASC5 | | Authors: | Petrovic, A, Mosalaganti, S, Keller, J, Mattiuzzo, M, Overlack, K, Wohlgemuth, S, Pasqualato, S, Raunser, S, Musacchio, A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modular Assembly of RWD Domains on the Mis12 Complex Underlies Outer Kinetochore Organization.
Mol.Cell, 53, 2014
|
|
1S0B
 
 | | Crystal structure of botulinum neurotoxin type B at pH 4.0 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
1S0D
 
 | | Crystal structure of botulinum neurotoxin type B at pH 5.5 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION, ZINC ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
1S0G
 
 | |
1S0C
 
 | | Crystal structure of botulinum neurotoxin type B at pH 5.0 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION, ZINC ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
5LSK
 
 | | CRYSTAL STRUCTURE OF THE HUMAN KINETOCHORE MIS12-CENP-C COMPLEX | | Descriptor: | Centromere protein C, Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, ... | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
5LSI
 
 | | CRYSTAL STRUCTURE OF THE KINETOCHORE MIS12 COMPLEX HEAD2 SUBDOMAIN CONTAINING DSN1 AND NSL1 FRAGMENTS | | Descriptor: | Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, SULFATE ION | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
5LSJ
 
 | | CRYSTAL STRUCTURE OF THE HUMAN KINETOCHORE MIS12-CENP-C delta-HEAD2 COMPLEX | | Descriptor: | Centromere protein C, Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, ... | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
4HV0
 
 | | Structure and Function of AvtR, a Novel Transcriptional Regulator from a Hyperthermophilic Archaeal Lipothrixvirus | | Descriptor: | AvtR | | Authors: | Peixeiro, N, Keller, J, Collinet, B, Leulliot, N, Campanacci, V, Cortez, D, Cambillau, C, Nitta, K.R, Vincentelli, R, Forterre, P, Prangishvili, D, Sezonov, G, van Tilbeurgh, H. | | Deposit date: | 2012-11-05 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Function of AvtR, a Novel Transcriptional Regulator from a Hyperthermophilic Archaeal Lipothrixvirus.
J.Virol., 87, 2013
|
|
1D7V
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH NMA | | Descriptor: | N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-2-METHYLALANINE, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1D7R
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH 5PA | | Descriptor: | N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1D7U
 
 | | Crystal structure of the complex of 2,2-dialkylglycine decarboxylase with LCS | | Descriptor: | POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), SODIUM ION, ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1D7S
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH DCS | | Descriptor: | D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
2WSI
 
 | |
2KEL
 
 | | Structure of the transcription regulator SvtR from the hyperthermophilic archaeal virus SIRV1 | | Descriptor: | Uncharacterized protein 56B | | Authors: | Guilliere, F, Kessler, A, Peixeiro, N, Sezonov, G, Prangishvili, D, Delepierre, M, Guijarro, J.I. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, function, and targets of the transcriptional regulator SvtR from the hyperthermophilic archaeal virus SIRV1.
J.Biol.Chem., 284, 2009
|
|
2IKQ
 
 | |
