8HVQ
 
 | | Crystal structure of haloacid dehalogenase-like hydrolase family enzyme from Staphylococcus lugdunensis | | Descriptor: | 1,2-ETHANEDIOL, Cof-type HAD-IIB family hydrolase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaur, H, Mahto, J.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Characterization of haloacid dehalogenase superfamily acid phosphatase from Staphylococcus lugdunensis.
Arch.Biochem.Biophys., 753, 2024
|
|
6QGX
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody F7 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoF7, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
6QGW
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody E6 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoE6, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
7NRF
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 2) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NRE
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 1) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NRI
 
 | | Structure of the darobactin-bound E. coli BAM complex (BamABCDE) | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
6QGY
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody B12 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoB12, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
6DN2
 
 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1354 SPLIT RNA | | Descriptor: | 4-{benzyl[2-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)ethyl]amino}butanoic acid, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
6DN3
 
 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1555 SPLIT RNA | | Descriptor: | 7,8-dimethyl-2,4-dioxo-10-(3-phenylpropyl)-1,2,3,4-tetrahydrobenzo[g]pteridin-10-ium, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
6DN1
 
 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1151 SPLIT RNA | | Descriptor: | 10-(6-carboxyhexyl)-8-(cyclopentylamino)-2,4-dihydroxy-7-methylbenzo[g]pteridin-10-ium, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
8FAZ
 
 | | Cryo-EM structure of the human BCDX2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair protein RAD51 homolog 2, DNA repair protein RAD51 homolog 3, ... | | Authors: | Jia, L, Wasmuth, E.V, Ruben, E.A, Sung, P, Rawal, Y, Greene, E.C, Meir, A, Olsen, S.K. | | Deposit date: | 2022-11-29 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural insights into BCDX2 complex function in homologous recombination.
Nature, 619, 2023
|
|
8QDU
 
 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*(FC)P*GP*(FC)P*GP*(FC)P*G)-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M.J. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
8QE4
 
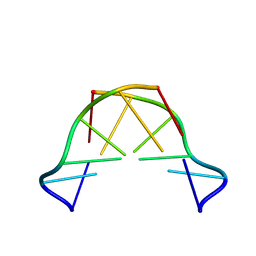 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*CP*(FRG)P*CP*(FRG)P*CP*(FRG))-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Friedland, D, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
8QOR
 
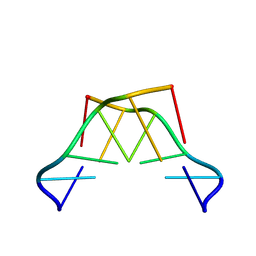 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*(CFL)P*(FRG)P*(CFL)P*(FRG)P*(CFL)P*(FRG))-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Friedland, D, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
8GBJ
 
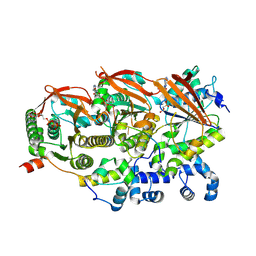 | | Cryo-EM structure of a human BCDX2/ssDNA complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*C)-3'), DNA repair protein RAD51 homolog 2, DNA repair protein RAD51 homolog 3, ... | | Authors: | Jia, L, Wasmuth, E.V, Ruben, E.A, Sung, P, Rawal, Y, Greene, E.C, Meir, A, Olsen, S.K. | | Deposit date: | 2023-02-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural insights into BCDX2 complex function in homologous recombination.
Nature, 619, 2023
|
|
4E1Q
 
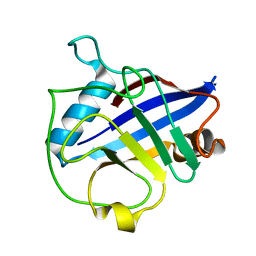 | | Crystal structure of Wheat Cyclophilin A at 1.25 A resolution | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Sekhon, S.S, Jeong, D.G, Woo, E.J, Singh, P, Yoon, T.S. | | Deposit date: | 2012-03-06 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structural and biochemical characterization of the cytosolic wheat cyclophilin TaCypA-1
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HY7
 
 | | Structural and biochemical characterization of a cytosolic wheat cyclophilin TaCypA-1 | | Descriptor: | Cyclosporin A, Peptidyl-prolyl cis-trans isomerase | | Authors: | Sekhon, S.S, Jeong, D.G, Woo, E.J, Singh, P, Pareek, A, Yoon, T.-S. | | Deposit date: | 2012-11-13 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and biochemical characterization of the cytosolic wheat cyclophilin TaCypA-1.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
