5HWI
 
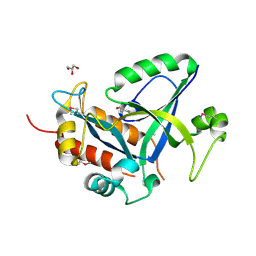 | | Crystal structure of selenomethionine labelled gama glutamyl cyclotransferease specific to glutathione from yeast | | Descriptor: | GLYCEROL, Glutathione-specific gamma-glutamylcyclotransferase, SUCCINIC ACID | | Authors: | Kaur, A, Gautam, R, Srivastava, R, Chandel, A, Kumar, A, Karthikeyan, S, Bachhawat, A.K. | | Deposit date: | 2016-01-29 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | ChaC2, an Enzyme for Slow Turnover of Cytosolic Glutathione
J. Biol. Chem., 292, 2017
|
|
5HWK
 
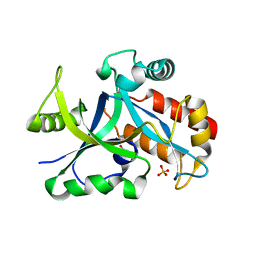 | | Crystal structure of gama glutamyl cyclotransferease specific to glutathione from yeast | | Descriptor: | BENZOIC ACID, Glutathione-specific gamma-glutamylcyclotransferase, PHOSPHATE ION | | Authors: | Kaur, A, Gautam, R, Srivastava, R, Chandel, A, Kumar, A, Karthikeyan, S, Bachhawat, A.K. | | Deposit date: | 2016-01-29 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.344 Å) | | Cite: | ChaC2, an Enzyme for Slow Turnover of Cytosolic Glutathione
J. Biol. Chem., 292, 2017
|
|
2RD4
 
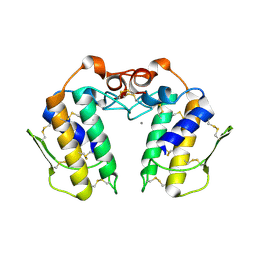 | | Design of specific inhibitors of Phospholipase A2: Crystal structure of the complex of phospholipase A2 with pentapeptide Leu-Val-Phe-Phe-Ala at 2.9 A resolution | | Descriptor: | CALCIUM ION, Phospholipase A2 isoform 1, Phospholipase A2 isoform 2, ... | | Authors: | Mirza, Z, Kaur, A, Singh, N, Sinha, M, Sharma, S, Srinivasan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-09-21 | | Release date: | 2007-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Design of specific inhibitors of Phospholipase A2: Crystal structure of the complex of phospholipase A2 with pentapeptide Leu-Val-Phe-Phe-Ala at 2.9 A resolution
To be Published
|
|
3CG9
 
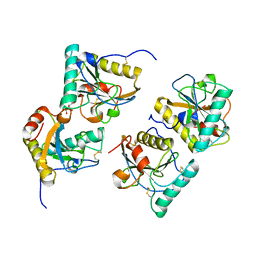 | | Crystal structure of the complex of peptidoglycan recognition protein with methyloxane-2,3,4,5-tetrol at 2.9 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein, alpha-L-rhamnopyranose | | Authors: | Sharma, P, Kaur, A, Singh, N, Sharma, S, Bhushan, A, Pathak, K.M.L, Kaur, P, Singh, T.P. | | Deposit date: | 2008-03-05 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein with methyoxane-2,3,4,5-tetrol at 2.9 A resolution
To be Published
|
|
3CR9
 
 | | Crystal structure of the complex of Lactoferrin with 6-(Hydroxymethyl)oxane-2,3,4,5-tetrol at 3.49 A resolution | | Descriptor: | FE (III) ION, Lactotransferrin, alpha-D-glucopyranose | | Authors: | Mir, R, Kaur, A, Singh, A.K, Singh, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2008-04-05 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal structure of the complex of Lactoferrin with 6-(Hydroxymethyl)oxane-2,3,4,5-tetrol at 3.49 A resolution
To be Published
|
|
8QC2
 
 | | Crystal structure of NAD-dependent glycoside hydrolase from Flavobacterium sp. (strain K172) in complex with co-factor NAD+ and sulfoquinovose (SQ) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Pickles, I.B, Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Widespread Family of NAD + -Dependent Sulfoquinovosidases at the Gateway to Sulfoquinovose Catabolism.
J.Am.Chem.Soc., 145, 2023
|
|
8QC3
 
 | |
8QC5
 
 | |
8QC8
 
 | |
8QC6
 
 | |
8OO2
 
 | | ChdA complex with amido-chelocardin | | Descriptor: | 2-carboxamido-2-deacetyl-chelocardin, MAGNESIUM ION, Putative transcriptional regulator | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2023-04-04 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Revision of the Absolute Configurations of Chelocardin and Amidochelocardin.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8V35
 
 | | Crystal structure of HpsN from Cupriavidus pinatubonensis | | Descriptor: | 1,2-ETHANEDIOL, Sulfopropanediol 3-dehydrogenase, ZINC ION | | Authors: | Lee, M. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and kinetic insights into the stereospecific oxidation of R -2,3-dihydroxypropanesulfonate by DHPS-3-dehydrogenase from Cupriavidus pinatubonensis.
Chem Sci, 15, 2024
|
|
8V37
 
 | |
8V36
 
 | |
9GYZ
 
 | |
9GYY
 
 | |
9CP8
 
 | |
9CP9
 
 | | Crystal structure of DHPS-3-dehydrogenase, HpsN H319A variant from Cupriavidus pinatubonensis in complex with substrate (R-DHPS) and NADH | | Descriptor: | (2R)-2,3-dihydroxypropane-1-sulfonic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Lee, M. | | Deposit date: | 2024-07-18 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Structural and kinetic insights into the stereospecific oxidation of R -2,3-dihydroxypropanesulfonate by DHPS-3-dehydrogenase from Cupriavidus pinatubonensis.
Chem Sci, 15, 2024
|
|
9CP7
 
 | |
8Q59
 
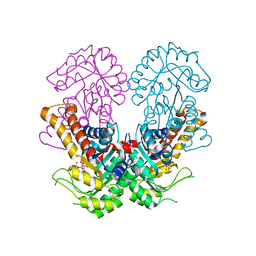 | | Crystal structure of metal-dependent class II sulfofructose phosphate aldolase from Yersinia aldovae in complex with sulfofructose phosphate (YaSqiA-Zn-SFP) | | Descriptor: | (3~{S},4~{S})-2,3,4-tris(oxidanyl)-5-oxidanylidene-6-phosphonooxy-hexane-1-sulfonic acid, SODIUM ION, Tagatose-1,6-bisphosphate aldolase kbaY, ... | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
8Q5A
 
 | | Crystal structure of metal-dependent class II sulfofructosephosphate aldolase from Hafnia paralvei HpSqiA-Zn in complex with dihydroxyacetone phosphate (DHAP) | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Ketose-bisphosphate aldolase, ZINC ION | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
8Q58
 
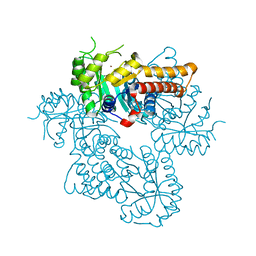 | |
8Q57
 
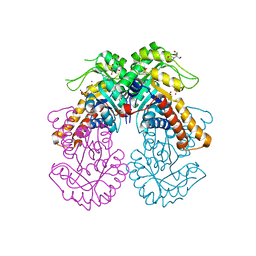 | | Crystal structure of class II SFP aldolase from Yersinia aldovae (YaSqiA-Zn-SO4) with bound sulfate ions | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, SULFATE ION, ... | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
5XXS
 
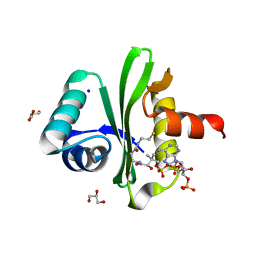 | |
5XXR
 
 | |
