5HWK
 
 | | Crystal structure of gama glutamyl cyclotransferease specific to glutathione from yeast | | Descriptor: | BENZOIC ACID, Glutathione-specific gamma-glutamylcyclotransferase, PHOSPHATE ION | | Authors: | Kaur, A, Gautam, R, Srivastava, R, Chandel, A, Kumar, A, Karthikeyan, S, Bachhawat, A.K. | | Deposit date: | 2016-01-29 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.344 Å) | | Cite: | ChaC2, an Enzyme for Slow Turnover of Cytosolic Glutathione
J. Biol. Chem., 292, 2017
|
|
5HWI
 
 | | Crystal structure of selenomethionine labelled gama glutamyl cyclotransferease specific to glutathione from yeast | | Descriptor: | GLYCEROL, Glutathione-specific gamma-glutamylcyclotransferase, SUCCINIC ACID | | Authors: | Kaur, A, Gautam, R, Srivastava, R, Chandel, A, Kumar, A, Karthikeyan, S, Bachhawat, A.K. | | Deposit date: | 2016-01-29 | | Release date: | 2016-12-14 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | ChaC2, an Enzyme for Slow Turnover of Cytosolic Glutathione
J. Biol. Chem., 292, 2017
|
|
5I2U
 
 | | Crystal structure of a novel Halo-Tolerant Cellulase from Soil Metagenome | | Descriptor: | Cellulase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Garg, R, Brahma, V, Srivastava, R, Verma, L, Karthikeyan, S, Sahni, G. | | Deposit date: | 2016-02-09 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural characterization of a novel halotolerant cellulase from soil metagenome
Sci Rep, 6, 2016
|
|
3TZ6
 
 | | Crystal structure of Aspartate semialdehyde dehydrogenase Complexed With inhibitor SMCS (CYS) And Phosphate From Mycobacterium tuberculosis H37Rv | | Descriptor: | Aspartate-semialdehyde dehydrogenase, CYSTEINE, GLYCEROL, ... | | Authors: | Vyas, R, Tewari, R, Weiss, M.S, Karthikeyan, S. | | Deposit date: | 2011-09-27 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of ternary complexes of aspartate-semialdehyde dehydrogenase (Rv3708c) from Mycobacterium tuberculosis H37Rv
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VOS
 
 | | Crystal structure of Aspartate semialdehyde dehydrogenase Complexed With glycerol and sulfate From Mycobacterium tuberculosis H37Rv | | Descriptor: | Aspartate-semialdehyde dehydrogenase, GLYCEROL, SULFATE ION | | Authors: | Vyas, R, Tewari, R, Weiss, M.S, Karthikeyan, S. | | Deposit date: | 2012-02-07 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of ternary complexes of aspartate-semialdehyde dehydrogenase (Rv3708c) from Mycobacterium tuberculosis H37Rv
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5YDC
 
 | |
4OW8
 
 | | Crystal structure of kinase domain of PknA from Mtb | | Descriptor: | GLYCEROL, SULFATE ION, Serine/threonine-protein kinase PknA | | Authors: | Ravala, S.K, Singh, S, Yadav, G.S, Karthikeyan, S, Chakraborti, P.K. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Evidence that phosphorylation of threonine in the GT motif triggers activation of PknA, a eukaryotic-type serine/threonine kinase from Mycobacterium tuberculosis.
Febs J., 282, 2015
|
|
4P8E
 
 | | Structure of ribB complexed with substrate (Ru5P) and metal ions | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P6C
 
 | | Structure of ribB complexed with inhibitor 4PEH | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P77
 
 | | Structure of ribB complexed with substrate Ru5P | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL, RIBULOSE-5-PHOSPHATE | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P6D
 
 | | Structure of ribB complexed with PO4 ion | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, PHOSPHATE ION | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P6P
 
 | | Structure of ribB complexed with inhibitor (4PEH) and metal ions | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID, ZINC ION | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P8J
 
 | | Structure of ribB | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
5XXS
 
 | |
5XXR
 
 | |
5Y06
 
 | |
5Y05
 
 | |
3LQU
 
 | |
3LRJ
 
 | |
3MIO
 
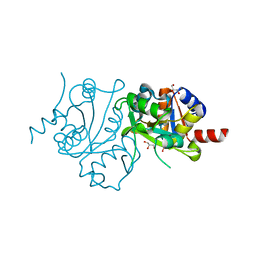 | | Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase domain from Mycobacterium tuberculosis at pH 6.00 | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Singh, M, Karthikeyan, S. | | Deposit date: | 2010-04-11 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for pH dependent monomer-dimer transition of 3,4-dihydroxy 2-butanone-4-phosphate synthase domain from Mycobacterium tuberculosis
J.Struct.Biol., 174, 2011
|
|
3MK5
 
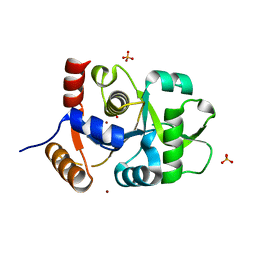 | | Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase domain from Mycobacterium tuberculosis with sulfate and zinc at pH 4.00 | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, SULFATE ION, ZINC ION | | Authors: | Singh, M, Karthikeyan, S. | | Deposit date: | 2010-04-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for pH dependent monomer-dimer transition of 3,4-dihydroxy 2-butanone-4-phosphate synthase domain from Mycobacterium tuberculosis
J.Struct.Biol., 174, 2011
|
|
3MGZ
 
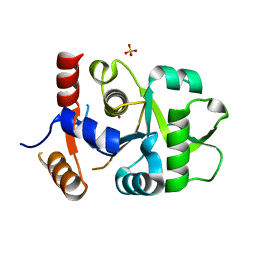 | | Crystal structure of DHBPS domain of bi-functional DHBPS/GTP cyclohydrolase II from Mycobacterium tuberculosis at pH 4.0 | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, SULFATE ION | | Authors: | Singh, M, Kumar, P, Karthikeyan, S. | | Deposit date: | 2010-04-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for pH dependent monomer-dimer transition of 3,4-dihydroxy 2-butanone-4-phosphate synthase domain from Mycobacterium tuberculosis
J.Struct.Biol., 174, 2011
|
|
3MK3
 
 | | Crystal structure of Lumazine synthase from Salmonella typhimurium LT2 | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, SULFATE ION | | Authors: | Kumar, P, Singh, M, Karthikeyan, S. | | Deposit date: | 2010-04-14 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.569 Å) | | Cite: | Crystal structure analysis of icosahedral lumazine synthase from Salmonella typhimurium, an antibacterial drug target.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3NQ4
 
 | |
1NUR
 
 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE | | Descriptor: | FKSG76, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
