5MF3
 
 | | NMR solution structure of Harzianin HK-VI in SDS micelles | | Descriptor: | Harzianin HK-VI | | Authors: | Kara, S, Zamora-Carreras, H, Afonin, S, Grage, S.L, Ulrich, A.S, Jimenez, M.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | 11-mer peptaibol Harzianin HK-VI: conformational and biological analysis
To Be Published
|
|
5MF8
 
 | | NMR solution structure of Harzianin HK-VI in trifluoroethanol | | Descriptor: | Harzianin HK-VI | | Authors: | Kara, S, Zamora-Carreras, H, Afonin, S, Grage, S.L, Ulrich, A.S, Jimenez, M.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | 11-mer peptaibol Harzianin HK-VI: conformational and biological analysis
To Be Published
|
|
5M9Y
 
 | | NMR solution structure of Harzianin HK-VI in DPC micelles | | Descriptor: | Harzianin HK-VI | | Authors: | Kara, S, Zamora-Carreras, H, Afonin, S, Grage, S.L, Ulrich, A.S, Jimenez, M.A. | | Deposit date: | 2016-11-02 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | 11-mer peptaibol Harzianin VI: conformational and biological analysis
To Be Published
|
|
3NB0
 
 | |
3O3C
 
 | | Glycogen synthase basal state UDP complex | | Descriptor: | Glycogen [starch] synthase isoform 2, SULFATE ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Baskaran, S, Hurley, T.D. | | Deposit date: | 2010-07-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.512 Å) | | Cite: | Structural basis for glucose-6-phosphate activation of glycogen synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NCH
 
 | |
7FCR
 
 | |
7FCS
 
 | |
3NAZ
 
 | | Basal state form of Yeast Glycogen Synthase | | Descriptor: | Glycogen [starch] synthase isoform 2, PEPTIDE, SULFATE ION | | Authors: | Baskaran, S, Hurley, T.D. | | Deposit date: | 2010-06-02 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for glucose-6-phosphate activation of glycogen synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4EXV
 
 | | Structure of Kluyveromyces lactis Hsv2p | | Descriptor: | SULFATE ION, SVP1-like protein 2 | | Authors: | Baskaran, S, Hurley, J.H. | | Deposit date: | 2012-05-01 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two-Site Recognition of Phosphatidylinositol 3-Phosphate by PROPPINs in Autophagy.
Mol.Cell, 47, 2012
|
|
3RT1
 
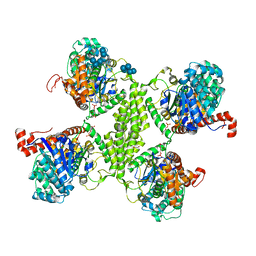 | |
3RSZ
 
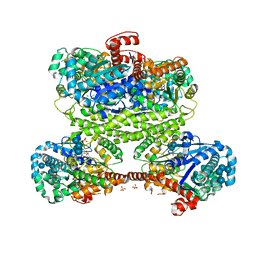 | | Maltodextran bound basal state conformation of yeast glycogen synthase isoform 2 | | Descriptor: | Glycogen [starch] synthase isoform 2, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Baskaran, S, Hurley, T.D. | | Deposit date: | 2011-05-02 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Multiple Glycogen-binding Sites in Eukaryotic Glycogen Synthase Are Required for High Catalytic Efficiency toward Glycogen.
J.Biol.Chem., 286, 2011
|
|
5SUK
 
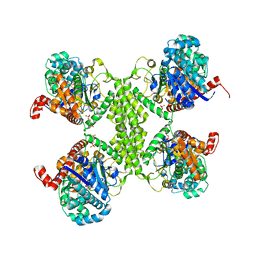 | |
4EC2
 
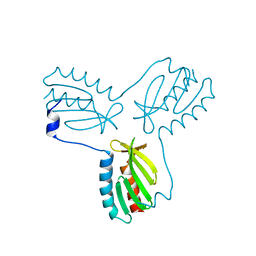 | | Crystal structure of trimeric frataxin from the yeast Saccharomyces cerevisiae, complexed with ferrous | | Descriptor: | FE (II) ION, Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2012-03-26 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The molecular basis of iron-induced oligomerization of frataxin and the role of the ferroxidation reaction in oligomerization.
J.Biol.Chem., 288, 2013
|
|
2FQL
 
 | |
7BNX
 
 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-22 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7BGS
 
 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-08 | | Release date: | 2022-01-19 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R4W
 
 | | Single stranded DNA binding protein SSB M5 from Fervidobacterium gondwanense | | Descriptor: | ACETIC ACID, PHOSPHATE ION, Single-stranded DNA-binding protein | | Authors: | Hakansson, M, Svensson, L.A, Werbowy, O, Al-Karadaghi, S, Kaczorowski, T, Kaczorowska, A.K, Dorawa, S. | | Deposit date: | 2022-02-09 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular characterization of a single stranded DNA binding protein from Fervidobacterium gondwanense
To Be Published
|
|
1LD3
 
 | | Crystal Structure of B. subilis ferrochelatase with Zn(2+) bound at the active site. | | Descriptor: | Ferrochelatase, ZINC ION | | Authors: | Lecerof, D, Fodje, M.N, Leon, R.A, Olsson, U, Hansson, A, Sigfridsson, E, Ryde, U, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-04-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Metal binding to Bacillus subtilis ferrochelatase and interaction between metal sites
J.Biol.Inorg.Chem., 8, 2003
|
|
1L8X
 
 | | Crystal Structure of Ferrochelatase from the Yeast, Saccharomyces cerevisiae, with Cobalt(II) as the Substrate Ion | | Descriptor: | COBALT (II) ION, Ferrochelatase | | Authors: | Karlberg, T, Lecerof, D, Gora, M, Silvegren, G, Labbe-Bois, R, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-03-22 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Metal Binding to Saccharomyces cerevisiae Ferrochelatase
Biochemistry, 41, 2002
|
|
2FOY
 
 | | Human Carbonic Anhydrase I complexed with a two-prong inhibitor | | Descriptor: | Carbonic anhydrase 1, ZINC ION, {2,2'-[(2-{[4-(AMINOSULFONYL)BENZOYL]AMINO}ETHYL)IMINO]DIACETATO(2-)-KAPPAO}COPPER | | Authors: | Jude, K.M, Banerjee, A.L, Haldar, M.K, Manokaran, S, Roy, B, Mallik, S, Srivastava, D.K, Christianson, D.W. | | Deposit date: | 2006-01-14 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ultrahigh resolution crystal structures of human carbonic anhydrases I and II complexed with two-prong inhibitors reveal the molecular basis of high affinity.
J.Am.Chem.Soc., 128, 2006
|
|
1AN7
 
 | |
7R0T
 
 | | Crystal structure of exonuclease ExnV1 | | Descriptor: | CHLORIDE ION, Exonuclease ExnV1, MAGNESIUM ION, ... | | Authors: | Welin, M, Svensson, A, Hakansson, M, Al-Karadaghi, S, Jasilionis, A, Linares-Pasten, J.A, Wang, L, Nordberg Karlsson, E, Ahlqvist, J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Crystal structure of DNA polymerase I from Thermus phage G20c.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R0K
 
 | | Crystal structure of Polymerase I from phage G20c | | Descriptor: | DNA polymerase I | | Authors: | Welin, M, Svensson, A, Hakansson, M, Al-Karadaghi, S, Linares-Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Ahlqvist, J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Crystal structure of DNA polymerase I from Thermus phage G20c.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1AD2
 
 | | RIBOSOMAL PROTEIN L1 MUTANT WITH SERINE 179 REPLACED BY CYSTEINE | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, MERCURY (II) ION, RIBOSOMAL PROTEIN L1, ... | | Authors: | Unge, J, Al-Karadaghi, S, Liljas, A, Jonsson, B.-H, Eliseikina, I, Ossina, N, Nevskaya, N, Fomenkova, N, Garber, M, Nikonov, S. | | Deposit date: | 1997-02-20 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A mutant form of the ribosomal protein L1 reveals conformational flexibility.
FEBS Lett., 411, 1997
|
|
