7NQS
 
 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with Z1203107138 | | Descriptor: | 1-ethyl-N-[(4-fluorophenyl)methyl]-1H-pyrazole-4-carboxamide, AMP PHOSPHORAMIDATE, GLYCEROL, ... | | Authors: | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | Deposit date: | 2021-03-02 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structures of P. falciparum Hsp70-x nucleotide binding domain with small molecule ligands
To Be Published
|
|
7OOG
 
 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with NCL-00023823 | | Descriptor: | 4-bromanylpyridin-2-amine, AMP PHOSPHORAMIDATE, CHLORIDE ION, ... | | Authors: | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | Deposit date: | 2021-05-27 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with NCL-00023823
To Be Published
|
|
7OOH
 
 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with NCL-00023818 | | Descriptor: | 4-IODOPYRAZOLE, AMP PHOSPHORAMIDATE, CHLORIDE ION, ... | | Authors: | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | Deposit date: | 2021-05-27 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with NCL-00023818
To Be Published
|
|
7OOE
 
 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with Z321318226 | | Descriptor: | AMP PHOSPHORAMIDATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | Deposit date: | 2021-05-27 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.369 Å) | | Cite: | Structures of P. falciparum Hsp70-x nucleotide binding domain with small molecule ligands
To Be Published
|
|
7P31
 
 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with NCL-00023818 | | Descriptor: | 4-IODOPYRAZOLE, AMP PHOSPHORAMIDATE, CHLORIDE ION, ... | | Authors: | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | Deposit date: | 2021-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with small molecule ligands
To Be Published
|
|
4MRA
 
 | | Crystal structure of Gpb in complex with QUERCETIN | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kantsadi, L.A, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2013-09-17 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Biochemical and biological assessment of the inhibitory potency of extracts from vinification byproducts of Vitis vinifera extracts against glycogen phosphorylase.
Food Chem.Toxicol., 67, 2014
|
|
6SSZ
 
 | | Structure of the Plasmodium falciparum falcipain 2 protease in complex with an (E)-chalcone inhibitor. | | Descriptor: | (~{E})-3-(1,3-benzodioxol-5-yl)-1-(3-nitrophenyl)prop-2-en-1-one, Cysteine proteinase falcipain 2a | | Authors: | Machin, J, Kantsadi, A, Vakonakis, I. | | Deposit date: | 2019-09-09 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The complex of Plasmodium falciparum falcipain-2 protease with an (E)-chalcone-based inhibitor highlights a novel, small, molecule-binding site.
Malar.J., 18, 2019
|
|
5OWY
 
 | | Glycogen Phosphorylase in complex with KS252 | | Descriptor: | 4-[5-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-4~{H}-1,2,4-triazol-3-yl]benzoic acid, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stravodimos, G, Kantsadi, A, Chatzileontiadou, D, Leonidas, D. | | Deposit date: | 2017-09-05 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the beta-pocket of the active site of human liver glycogen phosphorylase with 3-(C-beta-d-glucopyranosyl)-5-(4-substituted-phenyl)-1, 2, 4-triazole inhibitors.
Bioorg. Chem., 77, 2018
|
|
7ZKC
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum (apo form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Le Cornu, J.D. | | Deposit date: | 2022-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
7ZLU
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with UDP-2-deoxy-2-fluoro-D-glucose | | Descriptor: | 1,3-PROPANDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Ibba, R. | | Deposit date: | 2022-04-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
3SYM
 
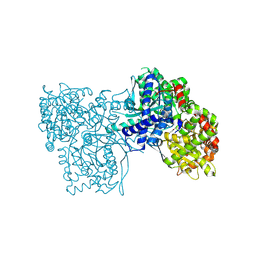 | | Glycogen Phosphorylase b in complex with 3 -C-(hydroxymethyl)-beta-D-glucopyranonucleoside of 5-fluorouracil | | Descriptor: | 5-fluoro-1-[3-C-(hydroxymethyl)-beta-D-glucopyranosyl]pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Skamnaki, V.T, Katsandi, A.L, Kontou, M, Leonidas, D.D. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3'-Axial CH(2) OH Substitution on Glucopyranose does not Increase Glycogen Phosphorylase Inhibitory Potency. QM/MM-PBSA Calculations Suggest Why.
Chem.Biol.Drug Des., 79, 2012
|
|
4YUA
 
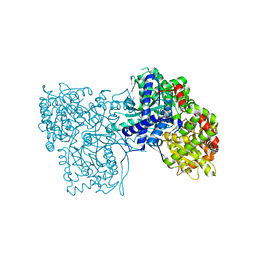 | | Glycogen phosphorylase in complex with ellagic acid | | Descriptor: | 2,3,7,8-tetrahydroxychromeno[5,4,3-cde]chromene-5,10-dione, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Kantsadi, L.A, Stravodimos, A.G, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2015-03-18 | | Release date: | 2015-05-13 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Natural flavonoids as antidiabetic agents. The binding of gallic and ellagic acids to glycogen phosphorylase b.
Febs Lett., 589, 2015
|
|
4Z5X
 
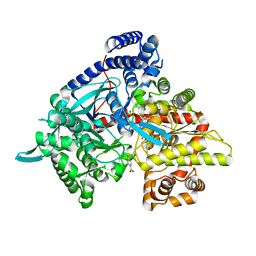 | | Glycogen phosphorylase in complex with gallic acid | | Descriptor: | 3,4,5-trihydroxybenzoic acid, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Kantsadi, L.A, Stravodimos, A.G, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2015-04-03 | | Release date: | 2015-05-13 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natural flavonoids as antidiabetic agents. The binding of gallic and ellagic acids to glycogen phosphorylase b.
Febs Lett., 589, 2015
|
|
4MI6
 
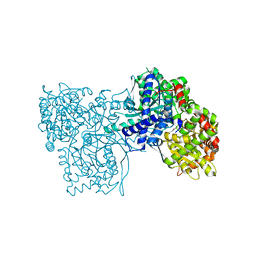 | | Crystal structure of Gpb in complex with SUGAR (N-[4-(5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)BUTANOYL]-BETA-D-GLUCOPYRANOSYLAMINE) | | Descriptor: | Glycogen phosphorylase, muscle form, N-[4-(5,6,7,8-tetrahydronaphthalen-2-yl)butanoyl]-beta-D-glucopyranosylamine | | Authors: | Kantsadi, L.A, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
4MHS
 
 | | Crystal structure of Gpb in complex with SUGAR (N-[(2E)-3-(BIPHENYL-4-YL)PROP-2-ENOYL]-BETA-D-GLUCOPYRANOSYLAMINE | | Descriptor: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Kantsadi, L.A, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
4MI3
 
 | | Crystal structure of Gpb in complex with SUGAR (N-{(2R)-2-METHYL-3-[4-(PROPAN-2-YL)PHENYL]PROPANOYL}-BETA-D-GLUCOPYRANOSYLAMINE) (S21) | | Descriptor: | Glycogen phosphorylase, muscle form, N-{(2R)-2-methyl-3-[4-(propan-2-yl)phenyl]propanoyl}-beta-D-glucopyranosylamine | | Authors: | Kantsadi, L.A, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
5EQO
 
 | |
5EPZ
 
 | |
5EOP
 
 | |
7GB5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-0da5ad92-2 (Mpro-x10201) | | Descriptor: | 2-(3-chlorophenyl)-N-(pyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.261 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GB3
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with DAV-CRI-3edb475e-6 (Mpro-x10172) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1S)-1-(3-chloro-5-fluorophenyl)ethyl]acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.379 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGZ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-3c65e9ce-2 (Mpro-x12719) | | Descriptor: | 2-(4-acetylpiperazin-1-yl)-N-(4-cyclopropylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBH
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-49816e9b-2 (Mpro-x10334) | | Descriptor: | 2-(3-chlorophenyl)-N-(2,4-dimethylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH0
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-ce760d3f-4 (Mpro-x12723) | | Descriptor: | (4R)-6-chloro-N-(2-oxo-2lambda~5~-isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBT
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BEN-DND-7e92b6ca-2 (Mpro-x10419) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[2-(2-methoxyphenoxy)ethyl]-N-methyl-2-oxo-1,2-dihydroquinoline-4-carboxamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
