5Z8B
 
 | |
1J1G
 
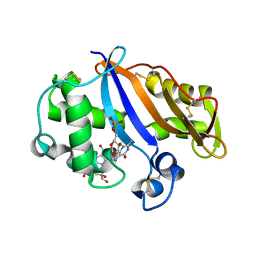 | | Crystal structure of the RNase MC1 mutant N71S in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Ribonuclease MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-04 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
1J1F
 
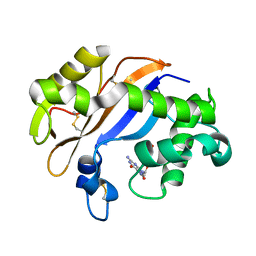 | | Crystal structure of the RNase MC1 mutant N71T in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
2ZOX
 
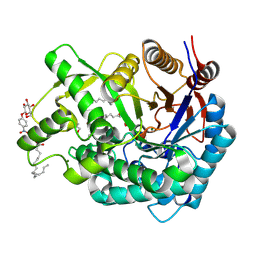 | | Crystal Structure of the Covalent Intermediate of Human Cytosolic beta-Glucosidase | | Descriptor: | 4-nitrophenyl alpha-D-glucopyranoside, Cytosolic beta-glucosidase, GLYCEROL, ... | | Authors: | Noguchi, J, Hayashi, Y, Baba, Y, Okino, N, Kimura, M, Ito, M, Kakuta, Y. | | Deposit date: | 2008-06-17 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the covalent intermediate of human cytosolic beta-glucosidase
Biochem.Biophys.Res.Commun., 374, 2008
|
|
2ZYT
 
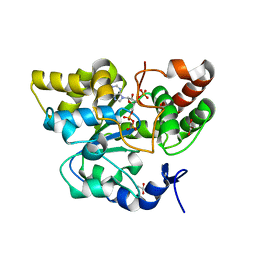 | | Crystal structure of mouse cytosolic sulfotransferase mSULT1D1 complex with PAPS | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, GLYCEROL, Tyrosine-ester sulfotransferase | | Authors: | Teramoto, T, Sakakibara, Y, Liu, M.-C, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Snapshot of a Michaelis complex in a sulfuryl transfer reaction: Crystal structure of a mouse sulfotransferase, mSULT1D1, complexed with donor substrate and accepter substrate
Biochem.Biophys.Res.Commun., 383, 2009
|
|
2ZXC
 
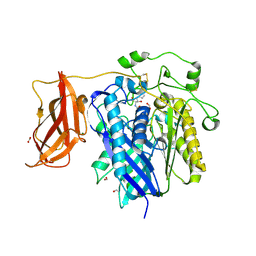 | | Ceramidase complexed with C2 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Okano, H, Inoue, T, Okino, N, Kakuta, Y, Matsumura, H, Ito, M. | | Deposit date: | 2008-12-22 | | Release date: | 2009-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase.
J.Biol.Chem., 284, 2009
|
|
3VKK
 
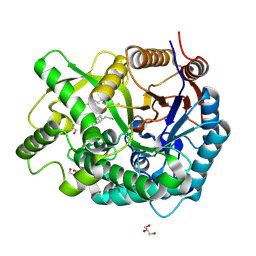 | | Crystal Structure Of The Covalent Intermediate Of Human Cytosolic Beta-Glucosidase-mannose complex | | Descriptor: | CHLORIDE ION, Cytosolic beta-glucosidase, GLYCEROL, ... | | Authors: | Noguchi, J, Hayashi, Y, Okino, N, Ito, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2011-11-17 | | Release date: | 2012-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition mechanism of human cytosolic beta-glucosidase by monnoside
To be Published
|
|
2CZW
 
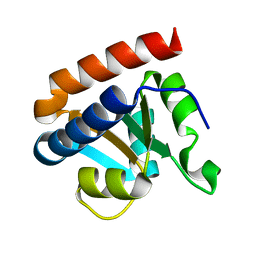 | | Crystal structure analysis of protein component Ph1496p of P.horikoshii ribonuclease P | | Descriptor: | 50S ribosomal protein L7Ae | | Authors: | Fukuhara, H, Kifusa, M, Watanabe, M, Terada, A, Honda, T, Numata, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A fifth protein subunit Ph1496p elevates the optimum temperature for the ribonuclease P activity from Pyrococcus horikoshii OT3
Biochem.Biophys.Res.Commun., 343, 2006
|
|
2CZV
 
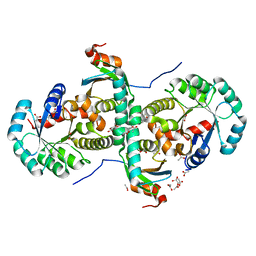 | | Crystal structure of archeal RNase P protein ph1481p in complex with ph1877p | | Descriptor: | ACETIC ACID, Ribonuclease P protein component 2, Ribonuclease P protein component 3, ... | | Authors: | Kawano, S, Kakuta, Y, Nakashima, T, Tanaka, I, Kimura, M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-06-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of protein Ph1481p in complex with protein Ph1877p of archaeal RNase P from Pyrococcus horikoshii OT3: implication of dimer formation of the holoenzyme
J.Mol.Biol., 357, 2006
|
|
1IYB
 
 | |
1UAR
 
 | |
7EH8
 
 | | Cryo-EM structure of the hexameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Minato, T, Teramoto, T, Adachi, N, Hung, N.K, Yamada, K, Kawasaki, M, Akutsu, M, Moriya, T, Senda, T, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
7EFV
 
 | | Crystal structure of octameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Minato, T, Teramoto, T, Hung, N.K, Yamada, K, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-23 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
7EFW
 
 | | Crystal structure of hexameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, GLYCEROL, ... | | Authors: | Minato, T, Teramoto, T, Hung, N.K, Yamada, K, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-23 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
7EH7
 
 | | Cryo-EM structure of the octameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Minato, T, Teramoto, T, Adachi, N, Hung, N.K, Yamada, K, Kawasaki, M, Akutsu, M, Moriya, T, Senda, T, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
2Z6V
 
 | | Crystal structure of sulfotransferase STF9 from Mycobacterium avium | | Descriptor: | PALMITIC ACID, Putative uncharacterized protein, SULFATE ION | | Authors: | Hassain, M.M, Moriizumi, Y, Tanaka, S, Kimura, M, Kakuta, Y. | | Deposit date: | 2007-08-09 | | Release date: | 2008-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of sulfotransferase STF9 from Mycobacterium avium
To be Published
|
|
2ZKC
 
 | | Crystal structure of human estrogen-related receptor gamma ligand binding domain complex with bisphenol Z | | Descriptor: | 4,4'-cyclohexane-1,1-diyldiphenol, Estrogen-related receptor gamma, GLYCEROL | | Authors: | Matsushima, A, Kakuta, Y, Teramoto, T, Shimohigashi, Y. | | Deposit date: | 2008-03-14 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human estrogen-related receptor gamma ligand binding domain complex with bisphenol Z
To be Published
|
|
2ZWI
 
 | | Crystal structure of alpha/beta-Galactoside alpha-2,3-Sialyltransferase from a Luminous Marine Bacterium, Photobacterium phosphoreum | | Descriptor: | Alpha-/beta-galactoside alpha-2,3-sialyltransferase, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Iwatani, T, Okino, N, Sakakura, M, Kajiwara, H, Ichikawa, M, Takakura, Y, Kimura, M, Ito, M, Yamamoto, T, Kakuta, Y. | | Deposit date: | 2008-12-05 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of alpha/beta-galactoside alpha2,3-sialyltransferase from a luminous marine bacterium, Photobacterium phosphoreum
Febs Lett., 583, 2009
|
|
2ZPT
 
 | | Crystal structure of mouse sulfotransferase SULT1D1 complex with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, GLYCEROL, Tyrosine-ester sulfotransferase | | Authors: | Teramoto, T, Sakakibara, Y, Inada, K, Liu, M.C, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2008-07-28 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of mSULT1D1, a mouse catecholamine sulfotransferase
Febs Lett., 582, 2008
|
|
2ZBS
 
 | |
2ZVQ
 
 | | Crystal structure of mouse cytosolic sulfotransferase mSULT1D1 complex with PAP and alpha-naphthol | | Descriptor: | 1-NAPHTHOL, ADENOSINE-3'-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Teramoto, T, Sakakibara, Y, Liu, M.-C, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the broad range substrate specificity of a novel mouse cytosolic sulfotransferase--mSULT1D1
Biochem.Biophys.Res.Commun., 379, 2009
|
|
2ZAS
 
 | | Crystal structure of human estrogen-related receptor gamma ligand binding domain complex with 4-alpha-cumylphenol, a bisphenol A derivative | | Descriptor: | 4-(1-methyl-1-phenylethyl)phenol, Estrogen-related receptor gamma, GLYCEROL | | Authors: | Matsushima, A, Kakuta, Y, Teramoto, T, Shimohigashi, Y. | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ERRgamma tethers strongly bisphenol A and 4-alpha-cumylphenol in an induced-fit manner
Biochem.Biophys.Res.Commun., 373, 2008
|
|
2ZVP
 
 | | Crystal structure of mouse cytosolic sulfotransferase mSULT1D1 complex with PAP and p-nitrophenol | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Teramoto, T, Sakakibara, Y, Liu, M.-C, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the broad range substrate specificity of a novel mouse cytosolic sulfotransferase--mSULT1D1
Biochem.Biophys.Res.Commun., 379, 2009
|
|
2ZYU
 
 | | Crystal structure of mouse cytosolic sulfotransferase mSULT1D1 complex with PAPS and p-nitrophenyl sulfate | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, 4-nitrophenyl sulfate, GLYCEROL, ... | | Authors: | Teramoto, T, Sakakibara, Y, Liu, M.-C, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Snapshot of a Michaelis complex in a sulfuryl transfer reaction: Crystal structure of a mouse sulfotransferase, mSULT1D1, complexed with donor substrate and accepter substrate
Biochem.Biophys.Res.Commun., 383, 2009
|
|
2ZYV
 
 | | Crystal structure of mouse cytosolic sulfotransferase mSULT1D1 complex with PAPS/PAP and p-nitrophenol | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Teramoto, T, Sakakibara, Y, Liu, M.-C, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Snapshot of a Michaelis complex in a sulfuryl transfer reaction: Crystal structure of a mouse sulfotransferase, mSULT1D1, complexed with donor substrate and accepter substrate
Biochem.Biophys.Res.Commun., 383, 2009
|
|
