1NST
 
 | |
1BO6
 
 | |
1AQU
 
 | |
1AQY
 
 | | ESTROGEN SULFOTRANSFERASE WITH PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, ESTROGEN SULFOTRANSFERASE | | Authors: | Kakuta, Y, Pedersen, L.G, Carter, C.W, Negishi, M, Pedersen, L.C. | | Deposit date: | 1997-08-04 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of estrogen sulphotransferase.
Nat.Struct.Biol., 4, 1997
|
|
1I7H
 
 | | CRYSTAL STURCUTURE OF FDX | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Kakuta, Y, Horio, T, Takahashi, Y, Fukuyama, K. | | Deposit date: | 2001-03-09 | | Release date: | 2002-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Escherichia coli Fdx, an adrenodoxin-type ferredoxin involved in the assembly of iron-sulfur clusters.
Biochemistry, 40, 2001
|
|
1X0U
 
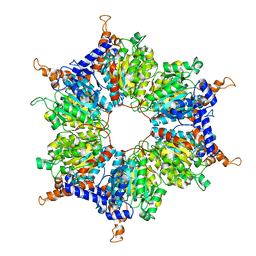 | |
1X0T
 
 | | Crystal structure of ribonuclease P protein Ph1601p from Pyrococcus horikoshii OT3 | | Descriptor: | Ribonuclease P protein component 4, ZINC ION | | Authors: | Kakuta, Y, Ishimatsu, I, Numata, T, Kimura, K, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2005-03-29 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Ribonuclease P Protein Ph1601p from Pyrococcus horikoshii OT3: An Archaeal Homologue of Human Nuclear Ribonuclease P Protein Rpp21(,)
Biochemistry, 44, 2005
|
|
1VB5
 
 | |
2Z4T
 
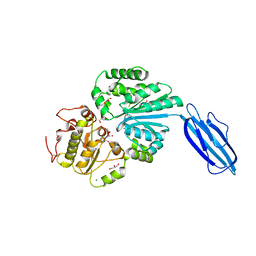 | | Crystal Structure of Vibrionaceae Photobacterium sp. JT-ISH-224 2,6-sialyltransferase in a Ternary Complex with Donor Product CMP and Accepter Substrate Lactose | | Descriptor: | Beta-galactoside alpha-2,6-sialyltransferase, CYTIDINE-5'-MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Kakuta, Y, Okino, N, Kajiwara, H, Ichikawa, M, Takakura, Y, Ito, M, Yamamoto, T. | | Deposit date: | 2007-06-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Vibrionaceae Photobacterium sp. JT-ISH-224 alpha2,6-sialyltransferase in a ternary complex with donor product CMP and acceptor substrate lactose: catalytic mechanism and substrate recognition
Glycobiology, 18, 2008
|
|
3AY8
 
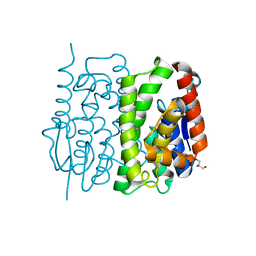 | | Glutathione S-transferase unclassified 2 from Bombyx mori | | Descriptor: | GLYCEROL, Glutathione S-transferase | | Authors: | Kakuta, Y, Usuda, K, Nakashima, T, Kimura, M, Aso, Y, Yamamoto, K. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic survey of active sites of an unclassified glutathione transferase from Bombyx mori
Biochim.Biophys.Acta, 1810, 2011
|
|
2E9L
 
 | | Crystal Structure of human Cytosolic Neutral beta-Glycosylceramidase (Klotho-related Prote:KLrP) complex with Glucose and fatty acids | | Descriptor: | Cytosolic beta-glucosidase, GLYCEROL, OLEIC ACID, ... | | Authors: | Kakuta, Y, Hayashi, Y, Okino, N, Ito, M. | | Deposit date: | 2007-01-25 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Klotho-related protein is a novel cytosolic neutral beta-glycosylceramidase.
J.Biol.Chem., 282, 2007
|
|
2E9M
 
 | | Crystal Structure of human Cytosolic Neutral beta-Glycosylceramidase (Klotho-related Prote:KLrP) complex with Galactose and fatty acids | | Descriptor: | Cytosolic beta-glucosidase, OLEIC ACID, PALMITIC ACID, ... | | Authors: | Kakuta, Y, Hayashi, Y, Okino, N, Ito, M. | | Deposit date: | 2007-01-25 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Klotho-related protein is a novel cytosolic neutral beta-glycosylceramidase.
J.Biol.Chem., 282, 2007
|
|
1UAI
 
 | |
3VK9
 
 | | Crystal structure of delta-class glutathione transferase from silkmoth | | Descriptor: | GLYCEROL, Glutathione S-transferase delta | | Authors: | Kakuta, Y, Usuda, K, Higashiura, A, Suzuki, M, Nakagawa, A, Kimura, M, Yamamoto, K. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for catalytic activity of a silkworm Delta-class glutathione transferase
Biochim.Biophys.Acta, 1820, 2012
|
|
2ZSC
 
 | | Tamavidin2, Novel Avidin-like Biotin-Binding Proteins from an Edible Mushroom | | Descriptor: | BIOTIN, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kakuta, Y, Okino, N, Ito, M, Yamamoto, T, Takakura, Y. | | Deposit date: | 2008-09-05 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Tamavidins--novel avidin-like biotin-binding proteins from the Tamogitake mushroom
Febs J., 276, 2009
|
|
2ZWS
 
 | | Crystal Structure Analysis of neutral ceramidase from Pseudomonas aeruginosa | | Descriptor: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kakuta, Y, Okino, N, Inoue, T, Okano, H, Ito, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase.
J.Biol.Chem., 284, 2009
|
|
3AMK
 
 | | Structure of the Starch Branching Enzyme I (BEI) from Oryza sativa L | | Descriptor: | GLYCEROL, Os06g0726400 protein, PHOSPHATE ION | | Authors: | Kakuta, Y, Chaen, K, Noguchi, J, Vu, N, Kimura, M. | | Deposit date: | 2010-08-20 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the branching enzyme I (BEI) from Oryza sativa L with implications for catalysis and substrate binding.
Glycobiology, 21, 2011
|
|
3AML
 
 | | Structure of the Starch Branching Enzyme I (BEI) from Oryza sativa L | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Kakuta, Y, Chaen, K, Noguchi, J, Vu, N, Kimura, M. | | Deposit date: | 2010-08-20 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the branching enzyme I (BEI) from Oryza sativa L with implications for catalysis and substrate binding.
Glycobiology, 21, 2011
|
|
8Y9P
 
 | |
5AX7
 
 | | yeast pyruvyltransferase Pvg1p | | Descriptor: | Pyruvyl transferase 1, ZINC ION | | Authors: | Kanekiyo, M, Yoritsune, K, Yoshinaga, S, Higuchi, Y, Takegawa, K, Kakuta, Y. | | Deposit date: | 2015-07-16 | | Release date: | 2016-06-08 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A rationally engineered yeast pyruvyltransferase Pvg1p introduces sialylation-like properties in neo-human-type complex oligosaccharide
Sci Rep, 6, 2016
|
|
8KDA
 
 | | Cryo-EM structure of Hydrogenobacter thermophilus minimal protein-only RNase P (HARP) in complex with pre-tRNAs | | Descriptor: | Aquifex aeolicus pre-tRNAVal, MAGNESIUM ION, RNA-free ribonuclease P | | Authors: | Teramoto, T, Adachi, N, Yokogawa, T, Koyasu, T, Mayanagi, K, Nakamura, T, Senda, T, Kakuta, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structure of Hydrogenobacter thermophilus minimal protein-only RNase P (HARP) in complex with pre-tRNAs
To Be Published
|
|
8KD9
 
 | | Cryo-EM structure of Aquifex aeolicus minimal protein-only RNase P (HARP) in complex with pre-tRNAs | | Descriptor: | Aquifex aeolicus pre-tRNAVal, RNA-free ribonuclease P | | Authors: | Teramoto, T, Koyasu, T, Mayanagi, K, Yokogawa, T, Adachi, N, Nakamura, T, Senda, T, Kakuta, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structure of Aquifex aeolicus minimal protein-only RNase P (HARP) in complex with pre-tRNAs
To Be Published
|
|
5WRJ
 
 | | Crystal structure of human tyrosylprotein sulfotransferase-1 complexed with PAP and gastrin peptide | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, MAGNESIUM ION, Protein-tyrosine sulfotransferase 1, ... | | Authors: | Tanaka, S, Nishiyori, T, Kojo, H, Otsubo, R, Kakuta, Y. | | Deposit date: | 2016-12-02 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the broad substrate specificity of the human tyrosylprotein sulfotransferase-1.
Sci Rep, 7, 2017
|
|
5WRI
 
 | | Crystal structure of human tyrosylprotein sulfotransferase-1 complexed with PAP and C4 peptide | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, ASP-PHE-GLU-ASP-TYR-GLU-PHE-ASP, GLYCEROL, ... | | Authors: | Tanaka, S, Nishiyori, T, Kojo, H, Otsubo, R, Kakuta, Y. | | Deposit date: | 2016-12-02 | | Release date: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the broad substrate specificity of the human tyrosylprotein sulfotransferase-1.
Sci Rep, 7, 2017
|
|
1DVG
 
 | | CRYSTAL STRUCTURE OF RAT HEME OXYGENASE-1 IN COMPLEX WITH HEME; SELELENO-METHIONINE DERIVATIVE, MUTATED AT M51T,M93L,M155L,M191L. | | Descriptor: | HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Omata, Y, Kakuta, Y, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2000-01-20 | | Release date: | 2000-04-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of rat heme oxygenase-1 in complex with heme.
FEBS Lett., 471, 2000
|
|
