2BUK
 
 | | SATELLITE TOBACCO NECROSIS VIRUS | | Descriptor: | CALCIUM ION, COAT PROTEIN | | Authors: | Jones, T.A, Liljas, L. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of Satellite Tobacco Necrosis Virus After Crystallographic Refinement at 2.5 A Resolution.
J.Mol.Biol., 177, 1984
|
|
8ADH
 
 | |
3CBH
 
 | |
1RBP
 
 | |
2A2G
 
 | | THE CRYSTAL STRUCTURES OF A2U-GLOBULIN AND ITS COMPLEX WITH A HYALINE DROPLET INDUCER. | | Descriptor: | D-LIMONENE 1,2-EPOXIDE, PROTEIN (ALPHA-2U-GLOBULIN) | | Authors: | Chaudhuri, B.N, Kleywegt, G.J, Jones, T.A. | | Deposit date: | 1998-11-19 | | Release date: | 1999-08-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structures of alpha 2u-globulin and its complex with a hyaline droplet inducer.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2A2U
 
 | |
2A39
 
 | | HUMICOLA INSOLENS ENDOCELLULASE EGI NATIVE STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Sulzenbacher, G, Mackenzie, L, Withers, S.G, Divne, C, Jones, T.A, Woldike, H.F, Schulein, M. | | Deposit date: | 1998-01-30 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate.
Biochem.J., 335, 1998
|
|
7CEL
 
 | | CBH1 (E217Q) IN COMPLEX WITH CELLOHEXAOSE AND CELLOBIOSE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
2A5V
 
 | | Crystal structure of M. tuberculosis beta carbonic anhydrase, Rv3588c, tetrameric form | | Descriptor: | CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE), THIOCYANATE ION, ZINC ION | | Authors: | Covarrubias, A.S, Bergfors, T, Jones, T.A, Hogbom, M. | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Mechanics of the pH-dependent Activity of beta-Carbonic Anhydrase from Mycobacterium tuberculosis
J.Biol.Chem., 281, 2006
|
|
1QK2
 
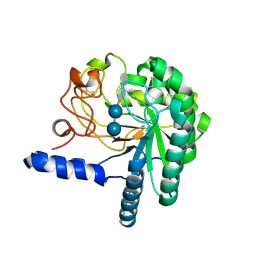 | | WILD TYPE CEL6A WITH A NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside, ... | | Authors: | Zou, J.-Y, Jones, T.A. | | Deposit date: | 1999-07-09 | | Release date: | 1999-09-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Evidence for Substrate Ring Distortion and Protein Conformational Changes During Catalysis in Cellobiohydrolase Cel6A from Trichoderma Reesei
Structure, 7, 1999
|
|
1QJW
 
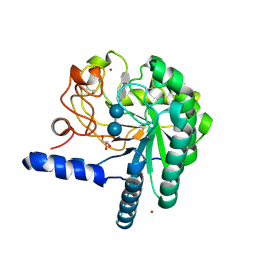 | | CEL6A (Y169F) WITH A NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), ... | | Authors: | Zou, J.-Y, Jones, T.A. | | Deposit date: | 1999-07-06 | | Release date: | 1999-09-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Evidence for Substrate Ring Distortion and Protein Conformational Changes During Catalysis in Cellobiohydrolase Cel6A from Trichoderma Reesei
Structure, 7, 1999
|
|
1HGY
 
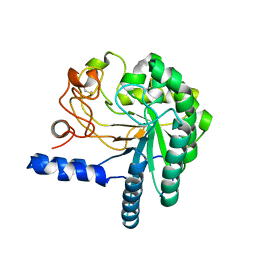 | | CEL6A D221A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), alpha-D-glucopyranose, ... | | Authors: | Zou, J.-Y, Kleywegt, G.J, Jones, T.A. | | Deposit date: | 2000-12-15 | | Release date: | 2002-01-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site of Cellobiohydrolase Cel6A from Trichoderma Reesei: The Roles of Aspartic Acids D221 and D175
J.Am.Chem.Soc., 124, 2002
|
|
1GSF
 
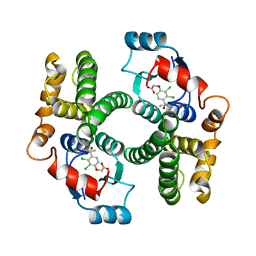 | | GLUTATHIONE TRANSFERASE A1-1 COMPLEXED WITH ETHACRYNIC ACID | | Descriptor: | ETHACRYNIC ACID, GLUTATHIONE TRANSFERASE A1-1 | | Authors: | L'Hermite, G, Sinning, I, Cameron, A.D, Jones, T.A. | | Deposit date: | 1995-06-09 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of human alpha-class glutathione transferase A1-1 in the apo-form and in complexes with ethacrynic acid and its glutathione conjugate.
Structure, 3, 1995
|
|
8QK5
 
 | | Structure of K. pneumoniae LpxH in complex with EBL-3647 | | Descriptor: | 5-[[3-(aminomethyl)azetidin-1-yl]methyl]-N-[4-[4-(4-cyano-6-methyl-pyrimidin-2-yl)piperazin-1-yl]sulfonylphenyl]-2-[methyl(methylsulfonyl)amino]benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QK2
 
 | | Structure of K.pneumoniae LpxH in complex with EBL-3339 | | Descriptor: | MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase, ~{N}-[4-[4-(4-cyano-6-methyl-pyrimidin-2-yl)piperazin-1-yl]sulfonylphenyl]-2-[methyl(methylsulfonyl)amino]benzamide | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QJZ
 
 | | Crystal structure of E. coli LpxH in complex with lipid X | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QKA
 
 | | Structure of K. pneumoniae LpxH in complex with JEDI-852 | | Descriptor: | 2-[methyl(methylsulfonyl)amino]-~{N}-(4-piperidin-1-ylsulfonylphenyl)benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QK9
 
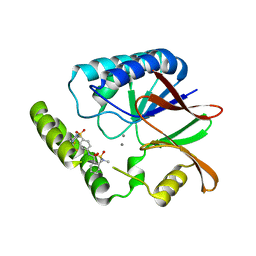 | | Structure of E. coli LpxH in complex with JEDI-1444 | | Descriptor: | 2-[methyl(methylsulfonyl)amino]-~{N}-[4-[4-[3-(trifluoromethyl)phenyl]piperazin-1-yl]sulfonylphenyl]benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6CEL
 
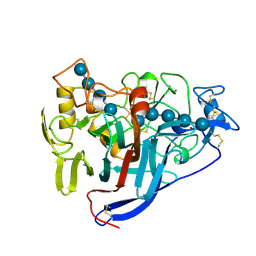 | | CBH1 (E212Q) CELLOPENTAOSE COMPLEX | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
5CEL
 
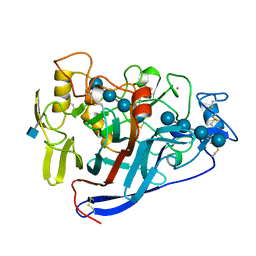 | | CBH1 (E212Q) CELLOTETRAOSE COMPLEX | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
1HB6
 
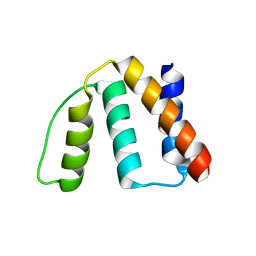 | | Structure of bovine Acyl-CoA binding protein in orthorhombic crystal form | | Descriptor: | ACYL-COA BINDING PROTEIN, CADMIUM ION | | Authors: | Zou, J.Y, Kleywegt, G.J, Bergfors, T, Knudsen, J, Jones, T.A. | | Deposit date: | 2001-04-12 | | Release date: | 2002-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding Site Differences Revealed by Crystal Structures of Plasmodium Falciparum and Bovine Acyl-Coa Binding Protein
J.Mol.Biol., 309, 2001
|
|
1H8V
 
 | | The X-ray Crystal Structure of the Trichoderma reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Shaw, A, Ropp, T.H, Wu, S, Bott, R, Cameron, A.D, Stahlberg, J, Mitchinson, C, Jones, T.A. | | Deposit date: | 2001-02-16 | | Release date: | 2001-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-Ray Crystal Structure of the Trichoderma Reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution
J.Mol.Biol., 308, 2001
|
|
1GUB
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | Descriptor: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, NICKEL (II) ION | | Authors: | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2002-01-24 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
1GUD
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | Descriptor: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, ZINC ION | | Authors: | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2002-01-24 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
1HB8
 
 | | Structure of bovine Acyl-CoA binding protein in tetragonal crystal form | | Descriptor: | ACYL-COA BINDING PROTEIN, SULFATE ION | | Authors: | Zou, J.Y, Kleywegt, G.J, Bergfors, T, Knudsen, J, Jones, T.A. | | Deposit date: | 2001-04-12 | | Release date: | 2002-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding Site Differences Revealed by Crystal Structures of Plasmodium Falciparum and Bovine Acyl-Coa Binding Protein
J.Mol.Biol., 309, 2001
|
|
