1VCA
 
 | | CRYSTAL STRUCTURE OF AN INTEGRIN-BINDING FRAGMENT OF VASCULAR CELL ADHESION MOLECULE-1 AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN VASCULAR CELL ADHESION MOLECULE-1 | | Authors: | Jones, E.Y, Harlos, K, Bottomley, M.J, Robinson, R.C, Driscoll, P.C, Edwards, R.M, Clements, J.M, Dudgeon, T.J, Stuart, D.I. | | Deposit date: | 1995-03-21 | | Release date: | 1995-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an integrin-binding fragment of vascular cell adhesion molecule-1 at 1.8 A resolution.
Nature, 373, 1995
|
|
1HNG
 
 | | CRYSTAL STRUCTURE AT 2.8 ANGSTROMS RESOLUTION OF A SOLUBLE FORM OF THE CELL ADHESION MOLECULE CD2 | | Descriptor: | CD2 | | Authors: | Jones, E.Y, Davis, S.J, Williams, A.F, Harlos, K, Stuart, D.I. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure at 2.8 A resolution of a soluble form of the cell adhesion molecule CD2.
Nature, 360, 1992
|
|
7B50
 
 | | Notum complex with ARUK3003778 | | Descriptor: | 3-(3,4-dichlorophenyl)sulfanyl-1$l^{4},2,7,8-tetrazabicyclo[4.3.0]nona-1(6),2,4,7-tetraen-9-one, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Jones, E.Y, Fish, P. | | Deposit date: | 2020-12-03 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
7B4X
 
 | | Notum complex with ARUK3002697 | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-methylphenyl)sulfanyl-2~{H}-[1,2,4]triazolo[4,3-b]pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Jones, E.Y, Fish, P. | | Deposit date: | 2020-12-02 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
4UYZ
 
 | |
6TFM
 
 | | Frizzled8 CRD | | Descriptor: | Frizzled-8 | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Antiepileptic Drug Carbamazepine Binds to a Novel Pocket on the Wnt Receptor Frizzled-8.
J.Med.Chem., 63, 2020
|
|
5FWV
 
 | | Wnt modulator Kremen crystal form III at 3.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, KREMEN PROTEIN 1 | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
5FWU
 
 | | Wnt modulator Kremen crystal form II at 2.8A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, KREMEN PROTEIN 1 | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
7B2Z
 
 | | Notum complex with ARUK3003907 | | Descriptor: | DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, SULFATE ION, ... | | Authors: | Zhao, Y, Jone, E.Y. | | Deposit date: | 2020-11-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural Insights into Notum Covalent Inhibition.
J.Med.Chem., 64, 2021
|
|
7B37
 
 | | Notum complex with ARUK3003718 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2,3-dihydroindol-1-yl)-4-oxidanylidene-butanoic acid, ... | | Authors: | Zhao, Y, Jone, E.Y. | | Deposit date: | 2020-11-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural Insights into Notum Covalent Inhibition.
J.Med.Chem., 64, 2021
|
|
7B2V
 
 | | Notum complex with ARUK3003906 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jone, E.Y. | | Deposit date: | 2020-11-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural Insights into Notum Covalent Inhibition.
J.Med.Chem., 64, 2021
|
|
7B2Y
 
 | | Notum complex with ARUK3003910 | | Descriptor: | 1,2-ETHANEDIOL, 2,2-bis(fluoranyl)ethyl 4-(2,3-dihydroindol-1-yl)-4-oxidanylidene-butanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jone, E.Y. | | Deposit date: | 2020-11-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural Insights into Notum Covalent Inhibition.
J.Med.Chem., 64, 2021
|
|
7B3F
 
 | | Notum S232A in complex with ARUK3003718 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2,3-dihydroindol-1-yl)-4-oxidanylidene-butanoic acid, ... | | Authors: | Zhao, Y, Jone, E.Y. | | Deposit date: | 2020-11-30 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Insights into Notum Covalent Inhibition.
J.Med.Chem., 64, 2021
|
|
7B3G
 
 | | Notum complex with ARUK3003902 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-((2-chlorophenyl)thio)-[1,2,4]triazolo[4,3-b]pyridazin-3(2H)-one, ... | | Authors: | Zhao, Y, Jone, E.Y. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
3GJF
 
 | | Rational development of high-affinity T-cell receptor-like antibodies | | Descriptor: | Antibody heavy chain, Antibody light chain, Beta-2-microglobulin, ... | | Authors: | Stewart-Jones, G, Wadle, A, Hombach, A, Shenderov, E, Held, G, Fischer, E, Kleber, S, Stenner-Liewen, F, Bauer, S, McMichael, A, Knuth, A, Abken, H, Hombach, A.A, Cerundolo, V, Jones, E.Y, Renner, C. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational development of high-affinity T-cell receptor-like antibodies
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GIV
 
 | | Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance | | Descriptor: | Beta-2-microglobulin, HIV-1 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Stewart-Jones, G, Iversen, A.K.N, Jones, E.Y. | | Deposit date: | 2009-03-06 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance
Nat.Immunol., 10, 2009
|
|
3GJE
 
 | | Rational development of high-affinity T-cell receptor-like antibodies | | Descriptor: | Fab Heavy Chain, Fab Light Chain | | Authors: | Stewart-Jones, G, Wadle, A, Hombach, A, Shenderov, E, Held, G, Fischer, E, Kleber, S, Stenner-Liewen, F, Bauer, S, McMichael, A, Knuth, A, Abken, H, Hombach, A.A, Cerundolo, V, Jones, E.Y, Renner, C. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational development of high-affinity T-cell receptor-like antibodies
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ZYI
 
 | | NetrinG2 in complex with NGL2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LEUCINE-RICH REPEAT-CONTAINING PROTEIN 4, ... | | Authors: | Seiradake, E, Coles, C.H, Perestenko, P.V, Harlos, K, McIlhinney, R.A.J, Aricescu, A.R, Jones, E.Y. | | Deposit date: | 2011-08-23 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Cell Surface Patterning Through Netring-Ngl Interactions
Embo J., 30, 2011
|
|
3ZYG
 
 | | NETRING2 LAM AND EGF1 DOMAINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NETRIN-G2 | | Authors: | Seiradake, E, Coles, C.H, Perestenko, P.V, Harlos, K, Mcilhinney, R.A.J, Aricescu, A.R, Jones, E.Y. | | Deposit date: | 2011-08-22 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Cell Surface Patterning Through Netring-Ngl Interactions.
Embo J., 30, 2011
|
|
3ZYO
 
 | | Crystal structure of the N-terminal leucine rich repeats and immunoglobulin domain of netrin-G ligand-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEUCINE-RICH REPEAT-CONTAINING PROTEIN 4B, ZINC ION | | Authors: | Seiradake, E, Coles, C.H, Perestenko, P.V, Harlos, K, McIlhinney, R.A.J, Aricescu, A.R, Jones, E.Y. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Cell Surface Patterning Through Netring-Ngl Interactions.
Embo J., 30, 2011
|
|
3ZYN
 
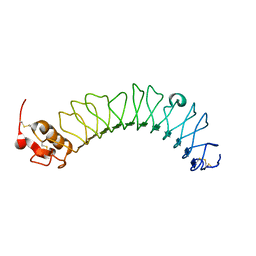 | | Crystal structure of the N-terminal leucine rich repeats of Netrin-G Ligand-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEUCINE-RICH REPEAT-CONTAINING PROTEIN 4B | | Authors: | Seiradake, E, Coles, C.H, Perestenko, P.V, Harlos, K, McIlhinney, R.A.J, Aricescu, A.R, Jones, E.Y. | | Deposit date: | 2011-08-23 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for Cell Surface Patterning Through Netring-Ngl Interactions.
Embo J., 30, 2011
|
|
3ZYJ
 
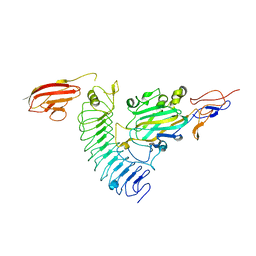 | | NetrinG1 in complex with NGL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LEUCINE-RICH REPEAT-CONTAINING PROTEIN 4C, ... | | Authors: | Seiradake, E, Coles, C.H, Perestenko, P.V, Harlos, K, McIlhinney, R.A.J, Aricescu, A.R, Jones, E.Y. | | Deposit date: | 2011-08-23 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis for Cell Surface Patterning Through Netring-Ngl Interactions.
Embo J., 30, 2011
|
|
7QVZ
 
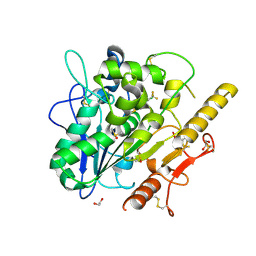 | | ARUK3001043_Notum | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Hillier, J, Willis, N.J, Mahy, W, Fish, P, Jones, E.Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure Activity Analysis of Notum Fragment Screen Hits with Development
To Be Published
|
|
2YPK
 
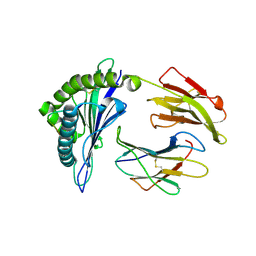 | | Structural features underlying T-cell receptor sensitivity to concealed MHC class I micropolymorphisms | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-57 ALPHA CHAIN, ... | | Authors: | Stewart-Jones, G.B, Simpson, P, Van Der Merwe, P.A, Easterbrook, P, Mcmichael, A.J, Rowland-Jones, S.L, Jones, E.Y, Gillespie, G.M. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Features Underlying T-Cell Receptor Sensitivity to Concealed Mhc Class I Micropolymorphisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6YUW
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 454 | | Descriptor: | 1-(cyclopropylmethyl)-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
