5NJ6
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in ternary complex with Fab3949 and AZ7188 at 4.0 angstrom resolution | | Descriptor: | Fab3949 H, Fab3949 L, Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2 | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
5NDD
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ8838 at 2.8 angstrom resolution | | Descriptor: | (~{S})-(4-fluoranyl-2-propyl-phenyl)-(1~{H}-imidazol-2-yl)methanol, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, PHOSPHATE ION, ... | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
5NDZ
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ3451 at 3.6 angstrom resolution | | Descriptor: | 2-(6-bromanyl-1,3-benzodioxol-5-yl)-~{N}-(4-cyanophenyl)-1-[(1~{S})-1-cyclohexylethyl]benzimidazole-5-carboxamide, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, SODIUM ION | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-09 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
4B78
 
 | | Aminoimidazoles as BACE-1 Inhibitors: From De Novo Design to Ab- lowering in Brain | | Descriptor: | (3R,5R)-3-methoxy-5-(4-methoxyphenyl)-5-(3-pyridin-3-ylphenyl)-3,4-dihydropyrrol-2-amine, BETA-SECRETASE 1 | | Authors: | Gravenfors, Y, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Berg, S, Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Rahm, F. | | Deposit date: | 2012-08-16 | | Release date: | 2013-06-26 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Core Refinement Toward Permeable Beta-Secretase (Bace-1) Inhibitors with Low Herg Activity.
J.Med.Chem., 56, 2013
|
|
4B72
 
 | | Aminoimidazoles as BACE-1 Inhibitors: From De Novo Design to Ab- lowering in Brain | | Descriptor: | (6R)-6-(4-methoxyphenyl)-2-methyl-6-(3-pyrimidin-5-ylphenyl)pyrrolo[3,4-d][1,3]thiazol-4-amine, BETA-SECRETASE 1 | | Authors: | Gravenfors, Y, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Berg, S, Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Rahm, F. | | Deposit date: | 2012-08-16 | | Release date: | 2013-06-26 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Core Refinement Toward Permeable Beta-Secretase (Bace-1) Inhibitors with Low Herg Activity.
J.Med.Chem., 56, 2013
|
|
4B70
 
 | | Aminoimidazoles as BACE-1 Inhibitors: From De Novo Design to Ab- lowering in Brain | | Descriptor: | (2S)-2-[3-(3-chlorophenyl)phenyl]-2-methyl-5,6-dihydro-1,3-oxazin-4-amine, BETA-SECRETASE 1 | | Authors: | Gravenfors, Y, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Berg, S, Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Rahm, F. | | Deposit date: | 2012-08-16 | | Release date: | 2013-06-26 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Core Refinement Toward Permeable Beta-Secretase (Bace-1) Inhibitors with Low Herg Activity.
J.Med.Chem., 56, 2013
|
|
4B77
 
 | | Aminoimidazoles as BACE-1 Inhibitors: From De Novo Design to Ab- lowering in Brain | | Descriptor: | (5R)-5-(4-methoxyphenyl)-5-(3-pyrimidin-5-ylphenyl)-3,4-dihydropyrrol-2-amine, BETA-SECRETASE 1, DIMETHYL SULFOXIDE | | Authors: | Gravenfors, Y, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Berg, S, Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Rahm, F. | | Deposit date: | 2012-08-16 | | Release date: | 2013-06-26 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Core Refinement Toward Permeable Beta-Secretase (Bace-1) Inhibitors with Low Herg Activity.
J.Med.Chem., 56, 2013
|
|
4AJM
 
 | | Development of a plate-based optical biosensor methodology to identify PDE10 fragment inhibitors | | Descriptor: | 3-AMINO-6-FLUORO-2-[4-(2-METHYLPYRIDIN-4-YL)PHENYL]-N-(METHYLSULFONYL)QUINOLINE-4-CARBOXAMIDE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Geschwindner, S, Johansson, P, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Albert, J.S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of a Plate-Based Optical Biosensor Methodology to Identify Pde10 Fragment Inhibitors
To be Published
|
|
1W74
 
 | | X-ray structure of peptidyl-prolyl cis-trans isomerase A, PpiA, Rv0009, from Mycobacterium tuberculosis. | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Henriksson, L.M, Johansson, P, Unge, T, Mowbray, S.L. | | Deposit date: | 2004-08-27 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure of Peptidyl-Prolyl Cis-Trans Isomerase a from Mycobacterium Tuberculosis
Eur.J.Biochem., 271, 2004
|
|
6QLZ
 
 | | IDOL F3ab subdomain | | Descriptor: | E3 ubiquitin-protein ligase MYLIP | | Authors: | Martinelli, L, Johansson, P, Wan, P.T, Gunnarsson, J, Guo, H, Boyd, H. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
2BI0
 
 | | RV0216, A conserved hypothetical protein from Mycobacterium tuberculosis that is essential for bacterial survival during infection, has a double hotdogfold | | Descriptor: | CHLORIDE ION, HYPOTHETICAL PROTEIN RV0216 | | Authors: | Castell, A, Johansson, P, Unge, T, Jones, T.A, Backbro, K. | | Deposit date: | 2005-01-20 | | Release date: | 2005-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rv0216, a Conserved Hypothetical Protein from Mycobacterium Tuberculosis that is Essential for Bacterial Survival During Infection, Has a Double Hotdog Fold
Protein Sci., 14, 2005
|
|
4AL0
 
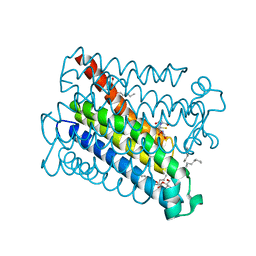 | | Crystal structure of Human PS-1 | | Descriptor: | GLUTATHIONE, PALMITIC ACID, PROSTAGLANDIN E SYNTHASE, ... | | Authors: | Sjogren, T, Nord, J, Ek, M, Johansson, P, Liu, G, Geschwindner, S. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal Structure of Microsomal Prostaglandin E2 Synthase Provides Insight Into Diversity in the Mapeg Superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AL1
 
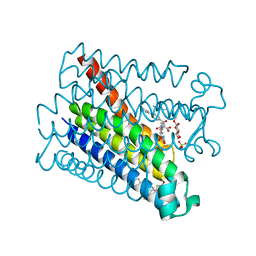 | | Crystal structure of Human PS-1 GSH-analog complex | | Descriptor: | L-gamma-glutamyl-S-(2-biphenyl-4-yl-2-oxoethyl)-L-cysteinylglycine, PALMITIC ACID, PROSTAGLANDIN E SYNTHASE, ... | | Authors: | Sjogren, T, Nord, J, Ek, M, Johansson, P, Liu, G, Geschwindner, S. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Microsomal Prostaglandin E2 Synthase Provides Insight Into Diversity in the Mapeg Superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4B1E
 
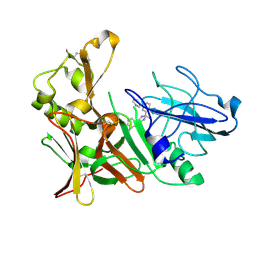 | | New Aminoimidazoles as BACE-1 Inhibitors: From Rational Design to Ab- lowering in Brain | | Descriptor: | (2R)-2-methyl-5-phenyl-2-(3-pyridin-3-ylphenyl)-2,3-dihydro-1H-imidazol-4-amine, BETA-SECRETASE 1 | | Authors: | Rahm, F, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, von Berg, S, von Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Gravenfors, Y. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-10 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New aminoimidazoles as beta-secretase (BACE-1) inhibitors showing amyloid-beta (A beta ) lowering in brain.
J. Med. Chem., 55, 2012
|
|
4B0Q
 
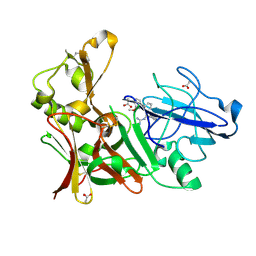 | |
4B1C
 
 | | New Aminoimidazoles as BACE-1 Inhibitors: From Rational Design to Ab- lowering in Brain | | Descriptor: | (2R)-2-cyclopropyl-5-methyl-2-[3-(5-prop-1-yn-1-ylpyridin-3-yl)phenyl]-2H-imidazol-4-amine, BETA-SECRETASE 1, DIMETHYL SULFOXIDE | | Authors: | Rahm, F, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, von Berg, S, von Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Gravenfors, Y. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-10 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New aminoimidazoles as beta-secretase (BACE-1) inhibitors showing amyloid-beta (A beta ) lowering in brain.
J. Med. Chem., 55, 2012
|
|
4B1D
 
 | | New Aminoimidazoles as BACE-1 Inhibitors: From Rational Design to Ab- lowering in Brain | | Descriptor: | (2S)-2-(4-methoxy-3,5-dimethylphenyl)-5-methyl-2-(3-pyrimidin-5-ylphenyl)-2H-imidazol-4-amine, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Rahm, F, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, von Berg, S, von Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Gravenfors, Y. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-10 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New aminoimidazoles as beta-secretase (BACE-1) inhibitors showing amyloid-beta (A beta ) lowering in brain.
J. Med. Chem., 55, 2012
|
|
2VEO
 
 | | X-ray structure of Candida antarctica lipase A in its closed state. | | Descriptor: | GLYCEROL, LIPASE A, TETRAETHYLENE GLYCOL, ... | | Authors: | Ericsson, D.J, Kasrayan, A, Johansson, P, Bergfors, T, Sandstrom, A.G, Backvall, J.E, Mowbray, S.L. | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-06 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Structure of Candida Antarctica Lipase a Shows a Novel Lid Structure and a Likely Mode of Interfacial Activation.
J.Mol.Biol., 376, 2008
|
|
2VX9
 
 | | H. salinarum dodecin E45A mutant | | Descriptor: | CHLORIDE ION, DODECIN, RIBOFLAVIN, ... | | Authors: | Grininger, M, Staudt, H, Johansson, P, Wachtveitl, J, Oesterhelt, D. | | Deposit date: | 2008-07-01 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dodecin is the Key Player in Flavin Homeostasis of Archaea.
J.Biol.Chem., 284, 2009
|
|
2VXA
 
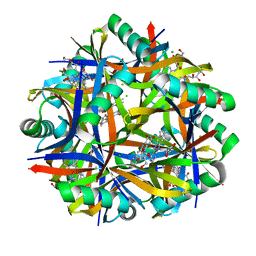 | | H. halophila dodecin in complex with riboflavin | | Descriptor: | CHLORIDE ION, DODECIN, RIBOFLAVIN | | Authors: | Grininger, M, Staudt, H, Johansson, P, Wachtveitl, J, Oesterhelt, D. | | Deposit date: | 2008-07-01 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dodecin is the Key Player in Flavin Homeostasis of Archaea.
J.Biol.Chem., 284, 2009
|
|
2WA1
 
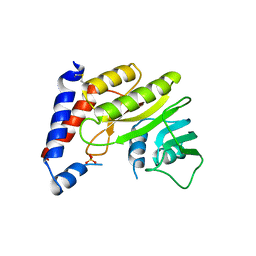 | | Structure of the methyltransferase domain from Modoc Virus, a Flavivirus with No Known Vector (NKV) | | Descriptor: | NON-STRUCTURAL PROTEIN 5, SULFATE ION | | Authors: | Jansson, A.M, Johansson, P, Jones, T.A. | | Deposit date: | 2009-02-02 | | Release date: | 2009-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Methyltransferase Domain from the Modoc Virus, a Flavivirus with No Known Vector.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2WA2
 
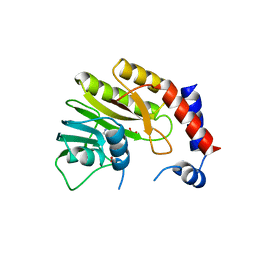 | | Structure of the methyltransferase domain from Modoc Virus, a Flavivirus with No Known Vector (NKV) | | Descriptor: | NON-STRUCTURAL PROTEIN 5, S-ADENOSYLMETHIONINE, SULFATE ION | | Authors: | Jansson, A.M, Johansson, P, Jones, T.A. | | Deposit date: | 2009-02-02 | | Release date: | 2009-08-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Methyltransferase Domain from the Modoc Virus, a Flavivirus with No Known Vector.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2X98
 
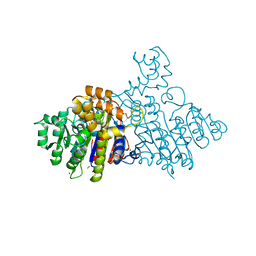 | | H.SALINARUM ALKALINE PHOSPHATASE | | Descriptor: | ALKALINE PHOSPHATASE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Wende, A, Johansson, P, Grininger, M, Oesterhelt, D. | | Deposit date: | 2010-03-14 | | Release date: | 2010-05-19 | | Last modified: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Characterization of a Halophilic Archaeal Alkaline Phosphatase.
J.Mol.Biol., 400, 2010
|
|
7PRV
 
 | | The glucocorticoid receptor in complex with fluticasone furoate, a PGC1a coactivator fragment and sgk 23bp | | Descriptor: | (6alpha,11alpha,14beta,16alpha,17alpha)-6,9-difluoro-17-{[(fluoromethyl)sulfanyl]carbonyl}-11-hydroxy-16-methyl-3-oxoan drosta-1,4-dien-17-yl furan-2-carboxylate, 1,2-ETHANEDIOL, DNA (5'-D(*TP*AP*CP*AP*GP*AP*AP*CP*AP*TP*TP*TP*TP*GP*TP*CP*CP*GP*TP*CP*GP*A)-3'), ... | | Authors: | Postel, S, Edman, K, Wissler, L. | | Deposit date: | 2021-09-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Quaternary glucocorticoid receptor structure highlights allosteric interdomain communication.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7PRX
 
 | | wildtype ligand binding domain of the glucocorticoid receptor complexed with velsecorat and a PGC1a coactivator fragment | | Descriptor: | Glucocorticoid receptor, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Velsecorat | | Authors: | Edman, K, Wissler, L, Koehler, C, Postel, S. | | Deposit date: | 2021-09-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Quaternary glucocorticoid receptor structure highlights allosteric interdomain communication.
Nat.Struct.Mol.Biol., 30, 2023
|
|
