4IZZ
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 21mer DNA target | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*AP*AP*CP*CP*CP*CP*TP*CP*AP*C)-3'), SULFATE ION, ... | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4J00
 
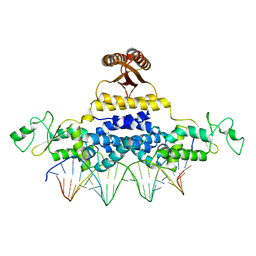 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 24mer DNA target | | Descriptor: | DNA (5'-D(*TP*GP*GP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*AP*AP*CP*CP*CP*CP*TP*CP*AP*CP*C)-3'), MAGNESIUM ION, Transcription Factor HetR | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1R8K
 
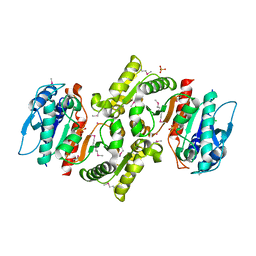 | | PDXA PROTEIN; NAD-DEPENDENT DEHYDROGENASE/CARBOXYLASE; SUBUNIT OF PYRIDOXINE PHOSPHATE BIOSYNTHETIC PROTEIN PDXJ-PDXA [SALMONELLA TYPHIMURIUM] | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase 1, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Osipiuk, J, Quartey, P, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-27 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of NAD-dependent dehydrogenase/carboxylase of Salmonella typhimurium
to be published
|
|
5HW2
 
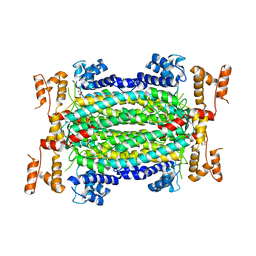 | | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with fumaric acid | | Descriptor: | 1,2-ETHANEDIOL, Adenylosuccinate lyase, FUMARIC ACID, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-28 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with fumaric acid
To Be Published
|
|
5HG0
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM | | Descriptor: | Pantothenate synthetase, S-ADENOSYLMETHIONINE | | Authors: | Chang, C, Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-07 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM
To Be Published
|
|
5HD6
 
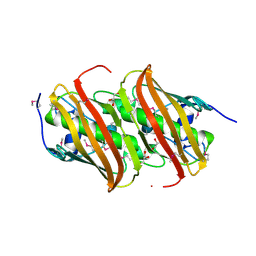 | | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A | | Descriptor: | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase, GLYCEROL | | Authors: | Chang, C, Maltseva, N, Kim, Y, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A
To Be Published
|
|
5HI6
 
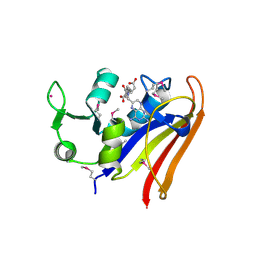 | | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-11 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form
To Be Published
|
|
5HKQ
 
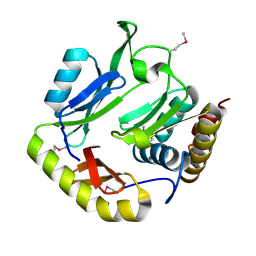 | | Crystal structure of CDI complex from Escherichia coli STEC_O31 | | Descriptor: | CdiI immunity protein, Contact-dependent inhibitor A | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional plasticity of antibacterial EndoU toxins.
Mol.Microbiol., 109, 2018
|
|
5HPF
 
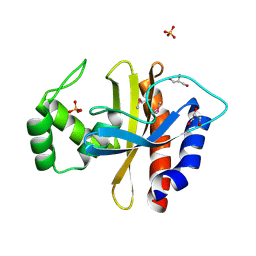 | | Crystal Structure of the Double Mutant of PobR Transcription Factor Inducer Binding Domain from Acinetobacter | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrejczak, R, Jha, R, Strauss, C.E.M, Joachimiak, A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | A microbial sensor for organophosphate hydrolysis exploiting an engineered specificity switch in a transcription factor.
Nucleic Acids Res., 44, 2016
|
|
4KQC
 
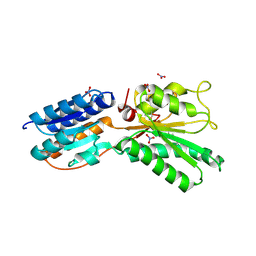 | | ABC transporter, LacI family transcriptional regulator from Brachyspira murdochii | | Descriptor: | NITRATE ION, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Osipiuk, J, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-14 | | Release date: | 2013-05-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | ABC transporter, LacI family transcriptional regulator from Brachyspira murdochii
To be Published
|
|
4KJM
 
 | | Crystal structure of the Staphylococcus aureus protein (NP_646141.1, domain 3912-4037) similar to streptococcal adhesins emb and ebhA/ebhB | | Descriptor: | ACETATE ION, CHLORIDE ION, Extracellular matrix-binding protein ebh, ... | | Authors: | Cymborowski, M, Shabalin, I.G, Joachimiak, G, Chruszcz, M, Gornicki, P, Zhang, R, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-03 | | Release date: | 2013-05-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Staphylococcus aureus protein (NP_646141.1, domain 3912-4037) similar to streptococcal adhesins emb and ebhA/ebhB
To be Published
|
|
5HJ5
 
 | | Crystal structure of tertiary complex of glucosamine-6-phosphate deaminase from Vibrio cholerae with BETA-D-GLUCOSE-6-PHOSPHATE and FRUCTOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, ACETIC ACID, FRUCTOSE -6-PHOSPHATE, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of tertiary complex of glucosamine-6-phosphate deaminase from Vibrio cholerae with BETA-D-GLUCOSE-6-PHOSPHATE and FRUCTOSE -6-PHOSPHATE
To Be Published
|
|
5HPI
 
 | | Crystal Structure of the Double Mutant of PobR Transcription Factor Inducer Binding Domain-3-Hydroxy Benzoic Acid complex from Acinetobacter | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYBENZOIC ACID, SULFATE ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrejczak, R, Jha, R, Strauss, C.E.M, Joachimiak, A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | A microbial sensor for organophosphate hydrolysis exploiting an engineered specificity switch in a transcription factor.
Nucleic Acids Res., 44, 2016
|
|
5HX7
 
 | | Metal ABC transporter from Listeria monocytogenes | | Descriptor: | Manganese-binding lipoprotein MntA | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-29 | | Release date: | 2016-02-17 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Metal ABC transporter from Listeria monocytogenes
to be published
|
|
5HVN
 
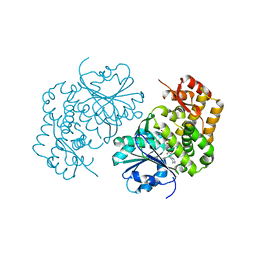 | | 3.0 Angstrom Crystal Structure of 3-dehydroquinate Synthase (AroB) from Francisella tularensis in Complex with NAD. | | Descriptor: | 3-dehydroquinate synthase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Dubrovska, I, Winsor, J, Zhou, M, Grimshaw, S, Kwon, K, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-28 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3.0 Angstrom Crystal Structure of 3-dehydroquinate Synthase (AroB) from Francisella tularensis in Complex with NAD.
To Be Published
|
|
1S3X
 
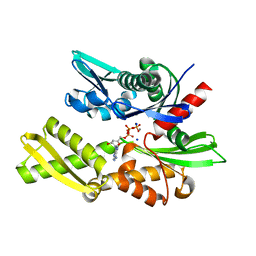 | | The crystal structure of the human Hsp70 ATPase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Heat shock 70 kDa protein 1, ... | | Authors: | Sriram, M, Osipiuk, J, Freeman, B, Morimoto, R.I, Joachimiak, A. | | Deposit date: | 2004-01-14 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Human Hsp70 molecular chaperone binds two calcium ions within the ATPase domain
Structure, 5, 1997
|
|
5HX0
 
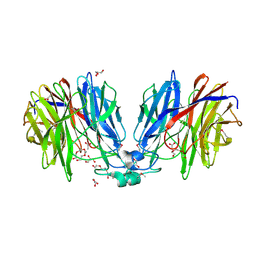 | | Crystal structure of unknown function protein Dfer_1899 fromDyadobacter fermentans DSM 18053 | | Descriptor: | ACETATE ION, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Chang, C, Duke, N, Clancy, S, Chhor, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-01-29 | | Release date: | 2016-02-17 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Crystal structure of unknown function protein Dfer_1899 fromDyadobacter fermentans DSM 18053
To Be Published
|
|
5I2H
 
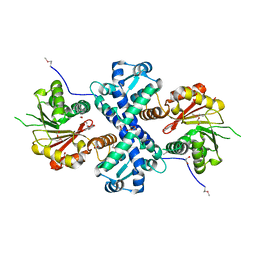 | | Crystal structure of O-methyltransferase family 2 protein Plim_1147 from Planctomyces limnophilus DSM 3776 complex with Apigenin | | Descriptor: | 1,2-ETHANEDIOL, 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, FORMIC ACID, ... | | Authors: | Chang, C, Duke, N, Bigelow, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-02-08 | | Release date: | 2016-03-02 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of O-methyltransferase family 2 protein Plim_1147 from Planctomyces limnophilus DSM 3776 complex with Apigenin.
To Be Published
|
|
5I47
 
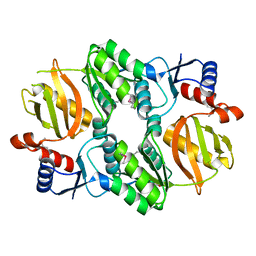 | | Crystal structure of RimK domain protein ATP-grasp from Sphaerobacter thermophilus DSM 20745 | | Descriptor: | GLYCEROL, RimK domain protein ATP-grasp | | Authors: | Chang, C, Duke, N, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-02-11 | | Release date: | 2016-03-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of RimK domain protein ATP-grasp from Sphaerobacter thermophilus DSM 20745
To Be Published
|
|
5I4K
 
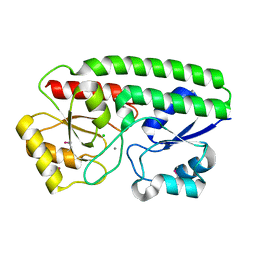 | | Metal ABC transporter from Listeria monocytogenes with manganese | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Manganese-binding lipoprotein MntA | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-12 | | Release date: | 2016-02-24 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Metal ABC transporter from Listeria monocytogenes with manganese
to be published
|
|
5I4Q
 
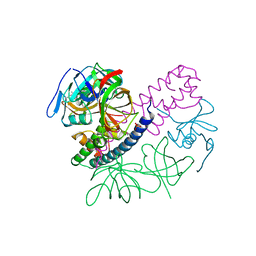 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (domains 2 and 3) | | Descriptor: | CHLORIDE ION, Contact-dependent inhibitor A, Contact-dependent inhibitor I, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2016-02-12 | | Release date: | 2017-06-28 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
1YLQ
 
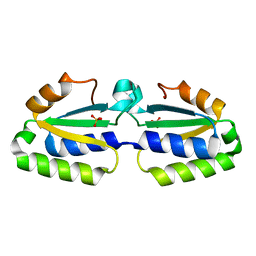 | | Crystal structure of putative nucleotidyltransferase | | Descriptor: | SULFATE ION, putative nucleotidyltransferase, hypothetical protein AF0614 | | Authors: | Chang, C, Joachimiak, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-19 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.016 Å) | | Cite: | Crystal structure of Hypothetical protein AF0614, putative nucleotidyltransferase
To be Published
|
|
5INT
 
 | | Crystal structure of the C-terminal Domain of Coenzyme A biosynthesis bifunctional protein CoaBC | | Descriptor: | Phosphopantothenate--cysteine ligase | | Authors: | Nocek, B, Zhou, M, Grimshaw, S, Kim, Y, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-07 | | Release date: | 2016-04-06 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the C-terminal Domain of Coenzyme A biosynthesis bifunctional protein CoaBC
To Be Published
|
|
5IR2
 
 | | Crystal structure of novel cellulases from microbes associated with the gut ecosystem | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Cellulase, ... | | Authors: | Chang, C, Mack, J, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-23 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Crystal structure of novel cellulases from microbes associated with the gut ecosystem
To Be Published
|
|
1YOY
 
 | |
