3LCB
 
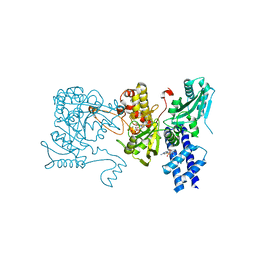 | | The crystal structure of isocitrate dehydrogenase kinase/phosphatase in complex with its substrate, isocitrate dehydrogenase, from Escherichia coli. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Isocitrate dehydrogenase [NADP], ... | | Authors: | Zheng, J, Jia, Z. | | Deposit date: | 2010-01-10 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the bifunctional isocitrate dehydrogenase kinase/phosphatase.
Nature, 465, 2010
|
|
3LMI
 
 | |
3LMH
 
 | |
3LC6
 
 | |
3LKM
 
 | |
3LLA
 
 | |
3KMH
 
 | |
3MPB
 
 | |
2MSI
 
 | |
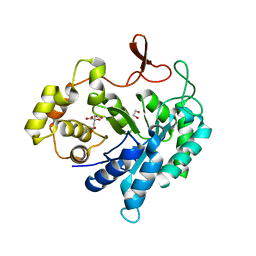
4GAC
 
 | |
4FL4
 
 | | Scaffoldin conformation and dynamics revealed by a ternary complex from the Clostridium thermocellum cellulosome | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Glycoside hydrolase family 9, ... | | Authors: | Currie, M.A, Adams, J.J, Faucher, F, Bayer, E.A, Jia, Z, Smith, S.P, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Scaffoldin Conformation and Dynamics Revealed by a Ternary Complex from the Clostridium thermocellum Cellulosome.
J.Biol.Chem., 287, 2012
|
|
4IL1
 
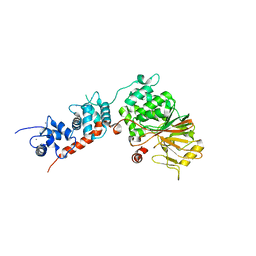 | | Crystal Structure of the Rat Calcineurin | | Descriptor: | CALCIUM ION, Calmodulin, Calcineurin subunit B type 1, ... | | Authors: | Ye, Q, Faucher, F, Jia, Z. | | Deposit date: | 2012-12-28 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of calcineurin activation by calmodulin.
Cell Signal, 25, 2013
|
|
4MF9
 
 | |
6IIH
 
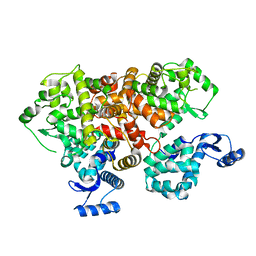 | | crystal structure of mitochondrial calcium uptake 2(MICU2) | | Descriptor: | CALCIUM ION, Endolysin,Calcium uptake protein 2, mitochondrial | | Authors: | Shen, Q, Wu, W, Zheng, J, Jia, Z. | | Deposit date: | 2018-10-06 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | The crystal structure of MICU2 provides insight into Ca2+binding and MICU1-MICU2 heterodimer formation.
Embo Rep., 20, 2019
|
|
2OVI
 
 | |
4MSI
 
 | |
6LB8
 
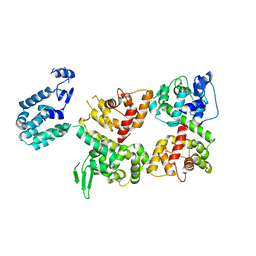 | | Crystal structure of the Ca2+-free T4L-MICU1-MICU2 complex | | Descriptor: | Calcium uptake protein 2, mitochondrial, Endolysin,Calcium uptake protein 1 | | Authors: | Wu, W, Shen, Q, Zheng, J, Jia, Z. | | Deposit date: | 2019-11-13 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | The structure of the MICU1-MICU2 complex unveils the regulation of the mitochondrial calcium uniporter.
Embo J., 39, 2020
|
|
6LB7
 
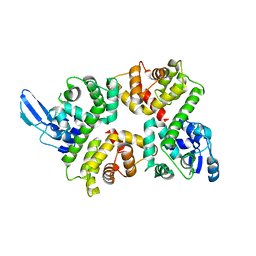 | | Crystal structure of the Ca2+-free and Ca2+-bound MICU1-MICU2 complex | | Descriptor: | CALCIUM ION, Calcium uptake protein 1, mitochondrial, ... | | Authors: | Wu, W, Shen, Q, Zheng, J, Jia, Z. | | Deposit date: | 2019-11-13 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | The structure of the MICU1-MICU2 complex unveils the regulation of the mitochondrial calcium uniporter.
Embo J., 39, 2020
|
|
3P2U
 
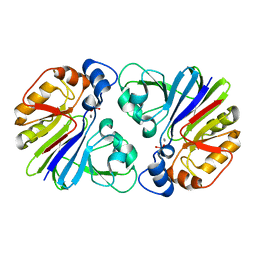 | | Crystal structure of PhnP in complex with orthovanadate | | Descriptor: | MANGANESE (II) ION, Protein phnP, VANADATE ION, ... | | Authors: | Podzelinska, K, Kim, Y.M, Zechel, D, Faucher, F, Jia, Z. | | Deposit date: | 2010-10-04 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of PhnP in complex with orthovanadate
TO BE PUBLISHED
|
|
3PDT
 
 | |
4MGF
 
 | |
3DXK
 
 | | Structure of Bos Taurus Arp2/3 Complex with Bound Inhibitor CK0944636 | | Descriptor: | Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, Actin-related protein 2/3 complex subunit 2, ... | | Authors: | Nolen, B.J, Tomasevic, N, Russell, A, Pierce, D.W, Jia, Z, Hartman, J, Sakowicz, R, Pollard, T.D. | | Deposit date: | 2008-07-24 | | Release date: | 2009-07-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization of two classes of small molecule inhibitors of Arp2/3 complex
Nature, 460, 2009
|
|
3CIO
 
 | |
3DRW
 
 | | Crystal Structure of a Phosphofructokinase from Pyrococcus horikoshii OT3 with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-specific phosphofructokinase, SODIUM ION | | Authors: | Singer, A.U, Skarina, T, Kochinyan, S, Brown, G, Cuff, M.E, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Jia, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-11 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ADP-dependent 6-phosphofructokinase from Pyrococcus horikoshii OT3: structure determination and biochemical characterization of PH1645.
J.Biol.Chem., 284, 2009
|
|
4P0U
 
 | |
