5NFY
 
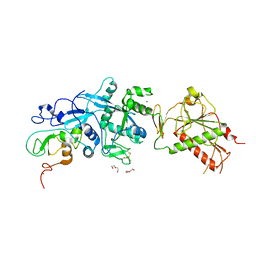 | | SARS-CoV nsp10/nsp14 dynamic complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, Polyprotein 1ab, ZINC ION | | Authors: | Ferron, F, Gluais, L, Vonrhein, C, Bricogne, G, Canard, B, Imbert, I. | | Deposit date: | 2017-03-16 | | Release date: | 2018-01-10 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (3.382 Å) | | Cite: | Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2XYV
 
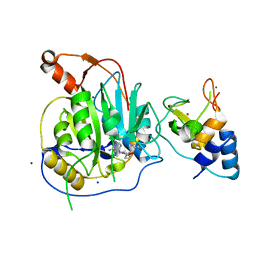 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-19 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2XYR
 
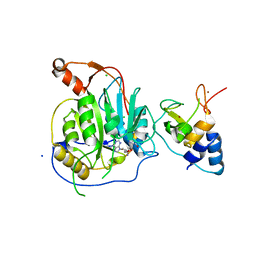 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2XYQ
 
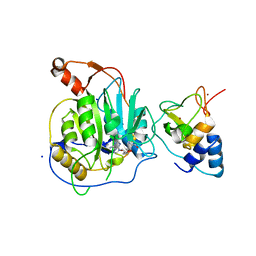 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2HL6
 
 | | Structure of homologously expressed Ferrulate esterase of Aspergillus niger in complex with CAPS | | Descriptor: | 1,2-ETHANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Feruloyl esterase A, ... | | Authors: | Parsiegla, G, Herpoel-Gimbert, I, Dubots, J. | | Deposit date: | 2006-07-06 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Respective importance of protein folding and glycosylation in the thermal stability of recombinant feruloyl esterase A
Febs Lett., 580, 2006
|
|
2XYB
 
 | | CRYSTAL STRUCTURE OF A FULLY FUNCTIONAL LACCASE FROM THE LIGNINOLYTIC FUNGUS PYCNOPORUS CINNABARINUS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Piontek, K, Choinowski, T, Antorini, M, Herpoel-Gimbert, I, Plattner, D.A, Sigoillot, J.C, Asther, M, Winterhalter, K. | | Deposit date: | 2010-11-17 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate Binding and Copper Geometry in Laccases
To be Published
|
|
7YZW
 
 | |
2L8K
 
 | | NMR Structure of the Arterivirus nonstructural protein 7 alpha (nsp7 alpha) | | Descriptor: | Non-structural protein 7 | | Authors: | Conte, M.R, Gaudin, C, Manolaridis, I, Tucker, P.W, Kelly, G. | | Deposit date: | 2011-01-19 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Genetic Analysis of the Arterivirus Nonstructural Protein 7{alpha}.
J.Virol., 85, 2011
|
|
5NO7
 
 | | Crystal Structure of a Xylan-active Lytic Polysaccharide Monooxygenase from Pycnoporus coccineus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lytic polysaccharide monooxygenase, SULFATE ION, ... | | Authors: | Ladeveze, S, Couturier, M, Sulzenbacher, G, Berrin, J.-G. | | Deposit date: | 2017-04-11 | | Release date: | 2018-01-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Lytic xylan oxidases from wood-decay fungi unlock biomass degradation.
Nat. Chem. Biol., 14, 2018
|
|
2IX9
 
 | |
1GYC
 
 | | CRYSTAL STRUCTURE DETERMINATION AT ROOM TEMPERATURE OF A LACCASE FROM TRAMETES VERSICOLOR IN ITS OXIDISED FORM CONTAINING A FULL COMPLEMENT OF COPPER IONS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Choinowski, T, Antorini, M, Piontek, K. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Laccase from the Fungus Trametes Versicolor at 1.90-A Resolution Containing a Full Complement of Coppers.
J.Biol.Chem., 277, 2002
|
|
1QKJ
 
 | | T4 Phage B-Glucosyltransferase, Substrate Binding and Proposed Catalytic Mechanism | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Imberty, I, Aschke-Sonnenborn, U, Ruger, W, Freemont, P.S. | | Deposit date: | 1999-07-22 | | Release date: | 1999-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | T4 Phage Beta-Glucosyltransferase: Substrate Binding and Proposed Catalytic Mechanism
J.Mol.Biol., 292, 1999
|
|
6FYK
 
 | | X-Ray structure of CLK2-KD(136-496)/Indazole1 at 2.39A | | Descriptor: | (2~{S})-4-methyl-1-[5-(3-methyl-2~{H}-indazol-5-yl)pyridin-3-yl]oxy-pentan-2-amine, Dual specificity protein kinase CLK2 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYL
 
 | | X-ray structure of CLK2-KD(136-496)/CX-4945 at 1.95A | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK2 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYO
 
 | | X-RAY STRUCTURE OF CLK1-KD(148-484)/Cpd-2 AT 2.32A | | Descriptor: | 6-~{tert}-butyl-~{N}-[6-(1~{H}-pyrazol-4-yl)-1~{H}-imidazo[1,2-a]pyridin-2-yl]pyridine-3-carboxamide, Dual specificity protein kinase CLK1, SULFATE ION | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYR
 
 | | X-RAY STRUCTURE OF CLK3-KD(GP-[275-632], NON-PHOS.)/Cpd-2 AT 1.42A | | Descriptor: | 6-~{tert}-butyl-~{N}-[6-(1~{H}-pyrazol-4-yl)-1~{H}-imidazo[1,2-a]pyridin-2-yl]pyridine-3-carboxamide, Dual specificity protein kinase CLK3 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYV
 
 | | X-RAY STRUCTURE OF CLK4-KD(146-480)/CX-4945 AT 2.46A | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK4, SULFATE ION | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYI
 
 | | X-ray Structure of CLK2-KD(130-496)/TG003 at 2.6A | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, Dual specificity protein kinase CLK2 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYP
 
 | | X-RAY STRUCTURE OF CLK3-KD(GP-[275-632], NON-PHOS.)/CX-4945 AT 2.29A | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK3 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
