5FJ4
 
 | |
3L72
 
 | | Chicken cytochrome BC1 complex with kresoxim-I-dimethyl bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, CYTOCHROME B, ... | | Authors: | Huang, L, Zhang, Z, Berry, E.A. | | Deposit date: | 2009-12-27 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Famoxadone and related inhibitors bind like methoxy acrylate inhibitors in the Qo site of the BC1 compl and fix the rieske iron-sulfur protein in a positio close to but distinct from that seen with stigmatellin and other "DISTAL" Qo inhibitors.
To be Published
|
|
3L70
 
 | | Cytochrome BC1 complex from chicken with trifloxystrobin bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Coenzyme Q10, ... | | Authors: | Huang, L, Berry, E.A. | | Deposit date: | 2009-12-27 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Famoxadone and related inhibitors bind like methoxy acrylate inhibitors in the Qo site of the BC1 compl and fix the rieske iron-sulfur protein in a positio close to but distinct from that seen with stigmatellin and other "distal" Qo inhibitors.
To be Published
|
|
3L75
 
 | | Cytochrome BC1 complex from chicken with fenamidone bound | | Descriptor: | (5S)-5-methyl-2-(methylsulfanyl)-5-phenyl-3-(phenylamino)-3,5-dihydro-4H-imidazol-4-one, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, AZIDE ION, ... | | Authors: | Huang, L, Berry, E.A. | | Deposit date: | 2009-12-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | FAMOXADONE AND RELATED INHIBITORS BIND LIKE METHOXY ACRYLATE INHIBITORS IN THE Qo SITE OF THE BC1 COMPL AND FIX THE RIESKE IRON-SULFUR PROTEIN IN A POSITIO CLOSE TO BUT DISTINCT FROM THAT SEEN WITH STIGMATELLIN AND OTHER "DISTAL" Qo INHIBITORS.
To be Published
|
|
3L71
 
 | | Cytochrome BC1 complex from chicken with azoxystrobin bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Coenzyme Q10, ... | | Authors: | Huang, L, Berry, E.A. | | Deposit date: | 2009-12-27 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Famoxadone and related inhibitors bind like methoxy acrylate inhibitors in the Qo site of the BC1 compl and fix the rieske iron-sulfur protein in a positio close to but distinct from that seen with stigmatellin and other "DISTAL" Qo inhibitors.
To be Published
|
|
3L74
 
 | | Cytochrome BC1 complex from chicken with famoxadone bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, AZIDE ION, CARDIOLIPIN, ... | | Authors: | Huang, L, Berry, E.A. | | Deposit date: | 2009-12-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Famoxadone and related inhibitors bind like methoxy acrylate inhibitors in the Qo site of the BC1 compl and fix the rieske iron-sulfur protein in a positio close to but distinct from that seen with stigmatellin and other "DISTAL" Qo inhibitors.
To be Published
|
|
3SUH
 
 | | Crystal structure of THF riboswitch, bound with 5-formyl-THF | | Descriptor: | N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, Riboswitch, SODIUM ION | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SUX
 
 | | Crystal structure of THF riboswitch, bound with THF | | Descriptor: | 5-HYDROXYMETHYLENE-6-HYDROFOLIC ACID, Riboswitch, SODIUM ION | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SUY
 
 | |
7EAG
 
 | | Crystal structure of the RAGATH-18 k-turn | | Descriptor: | RNA (5'-R(*GP*UP*CP*UP*AP*UP*GP*AP*AP*GP*GP*CP*UP*GP*GP*AP*GP*AP*C)-3') | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2021-03-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and folding of four putative kink turns identified in structured RNA species in a test of structural prediction rules.
Nucleic Acids Res., 49, 2021
|
|
7EAF
 
 | |
6HBT
 
 | |
6HBX
 
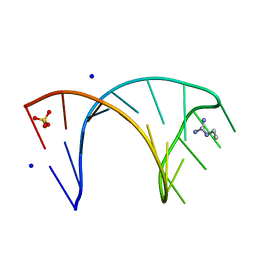 | |
6HCT
 
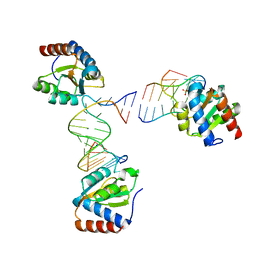 | |
6HC5
 
 | |
7XYC
 
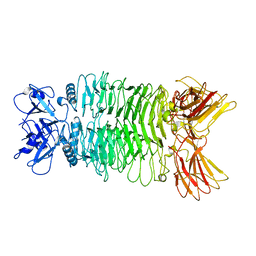 | |
7XY1
 
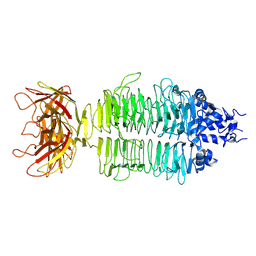 | |
7Y22
 
 | |
7Y3T
 
 | |
7Y1C
 
 | |
7Y23
 
 | |
7Y5S
 
 | |
1DUS
 
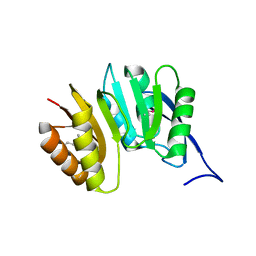 | | MJ0882-A hypothetical protein from M. jannaschii | | Descriptor: | MJ0882 | | Authors: | Hung, L, Huang, L, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-01-18 | | Release date: | 2000-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based experimental confirmation of biochemical function to a methyltransferase, MJ0882, from hyperthermophile Methanococcus jannaschii
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
3F3H
 
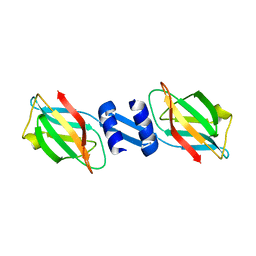 | | Crystal structure and anti-tumor activity of LZ-8 from the fungus Ganoderma lucidium | | Descriptor: | Immunomodulatory protein Ling Zhi-8 | | Authors: | Zhou, C.Z, Huang, L, He, Y.X, Bao, R, Sun, F, Liang, C, Liu, L. | | Deposit date: | 2008-10-30 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of LZ-8 from the medicinal fungus Ganoderma lucidium
Proteins, 75, 2009
|
|
6Q8U
 
 | |
