8KER
 
 | |
8KDT
 
 | |
8KDS
 
 | |
8KEP
 
 | |
8KEK
 
 | |
8KDM
 
 | |
8KDR
 
 | |
8KEJ
 
 | |
8KEO
 
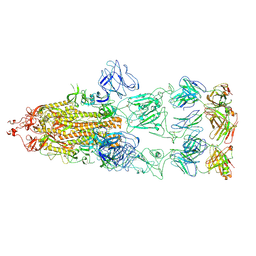 | |
5TKS
 
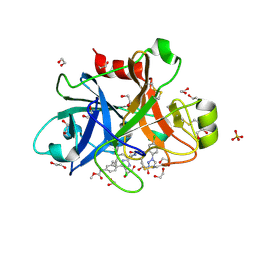 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR ((15S)-18-CHLORO- 15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2- PROPENOYL)AMINO)-17,19-DIAZATRICYCLO[14.2.1.0~2,7~]NONADECA-1(18),2,4,6,16(19)-PENTAEN-5-YL)CARBAMATE | | Descriptor: | ((15S)-18-CHLORO- 15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2- PROPENOYL)AMINO)-17,19-DIAZATRICYCLO[14.2.1.0~2,7~]NONADECA-1(18),2,4,6,16(19)-PENTAEN-5-YL)CARBAMATE, 1,2-ETHANEDIOL, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-10-07 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Based Design of Macrocyclic Factor XIa Inhibitors: Discovery of the Macrocyclic Amide Linker.
J. Med. Chem., 60, 2017
|
|
5TKU
 
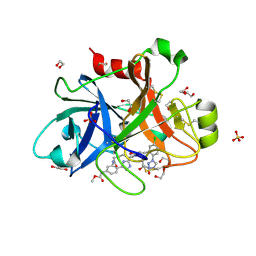 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR METHYL ((15S)-15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2-PROPENOYL)AMINO)-9-OXO-8,17,19-TRIAZATRICYCLO[14.2.1.0~2,7~]NONADECA-1(18),2,4,6,16(19)-PENTAEN-5-YL)CARBAMATE | | Descriptor: | 1,2-ETHANEDIOL, Factor XIa (Light Chain), METHYL ((15S)-15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2-PROPENOYL)AMINO)-9-OXO-8,17,19-TRIAZATRICYCLO[14.2.1.0~2,7~]N ONADECA-1(18),2,4,6,16(19)-PENTAEN-5-YL)CARBAMATE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-10-07 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Based Design of Macrocyclic Factor XIa Inhibitors: Discovery of the Macrocyclic Amide Linker.
J. Med. Chem., 60, 2017
|
|
5TKT
 
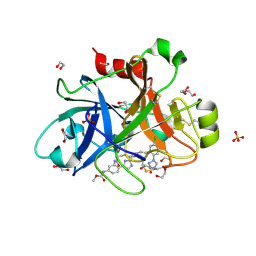 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR METHYL ((12E,15S)-15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2-PROPENOYL)AMINO)-9-OXO-8,17,19-TRIAZATRICYCLO[14.2.1.0~2,7~]NONADECA-1(18),2,4,6,12,16(19)-HEXAEN-5-YL)CARBAMATE | | Descriptor: | 1,2-ETHANEDIOL, Factor XIa (Light Chain), METHYL ((12E,15S)-15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2-PROPENOYL)AMINO)-9-OXO-8,17,19-TRIAZATRICYCLO[14.2.1.0~2, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-10-07 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Based Design of Macrocyclic Factor XIa Inhibitors: Discovery of the Macrocyclic Amide Linker.
J. Med. Chem., 60, 2017
|
|
5WSN
 
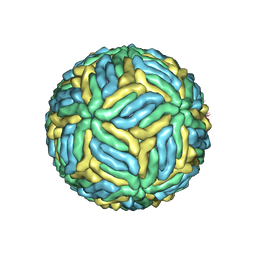 | | Structure of Japanese encephalitis virus | | Descriptor: | E protein, M protein | | Authors: | Wang, X, Zhu, L, Li, S, Yuan, S, Qin, C, Fry, E.E, Stuart, I.D, Rao, Z. | | Deposit date: | 2016-12-07 | | Release date: | 2017-05-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability
Nat Commun, 8, 2017
|
|
5U6I
 
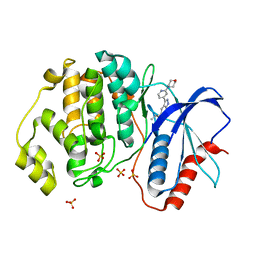 | | Discovery of MLi-2, an Orally Available and Selective LRRK2 Inhibitor that Reduces Brain Kinase Activity | | Descriptor: | 3-[2-(morpholin-4-yl)pyridin-4-yl]-5-[(propan-2-yl)oxy]-1H-indazole, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Scott, J.D, DeMong, D.E, Fell, M.J, Mirescu, C, Basu, K, Greshock, T.J, Morrow, J.A, Xiao, L, Hruza, A, Harris, J, Tiscia, H.E, Chang, R.K, Embrey, M.W, McCauley, J.A, Li, W, Lin, S, Liu, H, Dai, X, Baptista, M, Agnihotri, G, Columbus, J, Mei, H, Poirier, M, Zhou, X, Lin, Y, Yin, Z, Sanders, J.M, Drolet, R.E, Kern, J.T, Kennedy, M.E, Parker, E.M, Stamford, A.W, Nargund, R, Miller, M.W. | | Deposit date: | 2016-12-08 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of a 3-(4-Pyrimidinyl) Indazole (MLi-2), an Orally Available and Selective Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitor that Reduces Brain Kinase Activity.
J. Med. Chem., 60, 2017
|
|
3CEN
 
 | |
5EXN
 
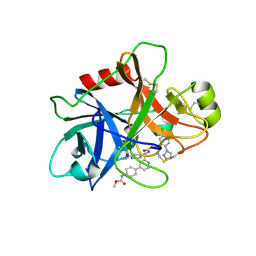 | | FACTOR XIA (C500S [C122S]) IN COMPLEX WITH THE INHIBITOR methyl ~{N}-[4-[2-[(1~{S})-1-[[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]amino]-2-phenyl-ethyl]pyridin-4-yl]phenyl]carbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, methyl ~{N}-[4-[2-[(1~{S})-1-[[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]amino]-2-phenyl-ethyl]pyridin-4-yl]phenyl]carbamate | | Authors: | Sheriff, S. | | Deposit date: | 2015-11-23 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Orally bioavailable pyridine and pyrimidine-based Factor XIa inhibitors: Discovery of the methyl N-phenyl carbamate P2 prime group
Bioorg.Med.Chem., 24, 2016
|
|
5EXL
 
 | |
5EXM
 
 | |
2P9C
 
 | |
2PA3
 
 | |
2P9G
 
 | |
2P9E
 
 | |
4LPH
 
 | | Crystal structure of human FPPS in complex with CL03093 | | Descriptor: | ({[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}methyl)phosphonic acid, Farnesyl pyrophosphate synthase, PHOSPHATE ION | | Authors: | Park, J, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-07-16 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multistage screening reveals chameleon ligands of the human farnesyl pyrophosphate synthase: implications to drug discovery for neurodegenerative diseases.
J.Med.Chem., 57, 2014
|
|
6KUZ
 
 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSL01 | | Descriptor: | 3-(1,3-benzothiazol-2-yl)-2-[[4-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]methoxy]-5-methyl-benzaldehyde, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Li, X.K, Guo, Y, Li, J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | First-generation species-selective chemical probes for fluorescence imaging of human senescence-associated beta-galactosidase.
Chem Sci, 11, 2020
|
|
7X1M
 
 | | The complex structure of Omicron BA.1 RBD with BD604, S309,and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab heavy chain, BD-604 Fab light chain, ... | | Authors: | Huang, M, Xie, Y.F, Qi, J.X. | | Deposit date: | 2022-02-24 | | Release date: | 2022-07-06 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3.
Immunity, 55, 2022
|
|
