9CJ0
 
 | |
4XD1
 
 | | X-ray structure of the N-formyltransferase QdtF from Providencia alcalifaciens, W305A mutant, in the presence of TDP-Qui3N and N5-THF | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Thoden, J.B, Woodford, C.R, Holden, H.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New Role for the Ankyrin Repeat Revealed by a Study of the N-Formyltransferase from Providencia alcalifaciens.
Biochemistry, 54, 2015
|
|
3MQG
 
 | | crystal structure of the 3-N-acetyl transferase WlbB from Bordetella petrii in complex with acetyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ACETYL COENZYME *A, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2010-04-28 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Molecular structure of WlbB, a bacterial N-acetyltransferase involved in the biosynthesis of 2,3-diacetamido-2,3-dideoxy-D-mannuronic acid .
Biochemistry, 49, 2010
|
|
4XD0
 
 | | X-ray structure of the N-formyltransferase QdtF from Providencia alcalifaciens | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, CHLORIDE ION, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Thoden, J.B, Woodford, C.R, Holden, H.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New Role for the Ankyrin Repeat Revealed by a Study of the N-Formyltransferase from Providencia alcalifaciens.
Biochemistry, 54, 2015
|
|
3MQH
 
 | | crystal structure of the 3-N-acetyl transferase WlbB from Bordetella petrii in complex with CoA and UDP-3-amino-2-acetamido-2,3-dideoxy glucuronic acid | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-4-amino-6-{[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxytetrahydro-2H-pyran-2-carboxylic acid, 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | thoden, J.B, holden, H.M. | | Deposit date: | 2010-04-28 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Molecular structure of WlbB, a bacterial N-acetyltransferase involved in the biosynthesis of 2,3-diacetamido-2,3-dideoxy-D-mannuronic acid .
Biochemistry, 49, 2010
|
|
3M9V
 
 | |
5TIF
 
 | | x-ray structure of acyl-CoA thioesterase I, TesA, triple mutant M141L/Y145K/L146K in complex with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, DI(HYDROXYETHYL)ETHER, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
5TIE
 
 | | x-ray structure of acyl-CoA thioesterase I, TesA, mutant M141L/Y145K/L146K at pH 7.5 in complex with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
7TXS
 
 | | X-ray structure of the VioB N-aetyltransferase from Acinetobacter baumannii in the presence of a reaction intermediate | | Descriptor: | SODIUM ION, VioB, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methyl (3R)-4-({3-[(2-{[(1S)-1-{[(2R,3S,4S,5R,6R)-4,5-dihydroxy-6-{[(R)-hydroxy{[(R)-hydroxy{[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)oxolan-2-yl]methoxy}phosphoryl]oxy}phosphoryl]oxy}-2-methyloxan-3-yl]amino}ethyl]sulfanyl}ethyl)amino]-3-oxopropyl}amino)-3-hydroxy-2,2-dimethyl-4-oxobutyl dihydrogen diphosphate (non-preferred name) | | Authors: | Herkert, N.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure and function of an N-acetyltransferase from the human pathogen Acinetobacter baumannii isolate BAL_212.
Proteins, 90, 2022
|
|
7TXQ
 
 | | x-ray structure of the VioB N-acetyltransferase from Acinetobacter baumannii in the present of TDP and Acetyl-CoenzymeA | | Descriptor: | ACETYL COENZYME *A, SODIUM ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Herkert, N.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and function of an N-acetyltransferase from the human pathogen Acinetobacter baumannii isolate BAL_212.
Proteins, 90, 2022
|
|
7TXP
 
 | | X-ray structure of the VioB N-acetyltransferase from Acinetobacter baumannii in complex with TDP-4-amino-4,6-dideoxy-D-glucose | | Descriptor: | SODIUM ION, VioB, dTDP-4-amino-4,6-dideoxyglucose | | Authors: | Herkert, N.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and function of an N-acetyltransferase from the human pathogen Acinetobacter baumannii isolate BAL_212.
Proteins, 90, 2022
|
|
7US5
 
 | | X-ray crystal structure of GDP-D-glycero-D-manno-heptose 4,6-Dehydratase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-GLYCERO-D-MANNO-HEPTOSE 4,6-DEHYDRATASE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-04-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction Mechanism and Three-Dimensional Structure of GDP-d-glycero-alpha-d-manno-heptose 4,6-Dehydratase from Campylobacter jejuni.
Biochemistry, 61, 2022
|
|
2UDP
 
 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHENYL-URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray structure of UDP-galactose 4-epimerase complexed with UDP-phenol.
Protein Sci., 5, 1996
|
|
8E77
 
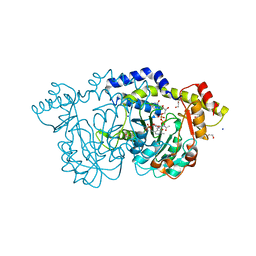 | | rystal structure of Pcryo_0616, the aminotransferase required to synthesize UDP-N-acetyl-3-amino-D-glucosaminuronic acid (UDP-GlcNAc3NA), incomplete with its external aldimine reaction intermediate | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-6-{[(R)-{[(S)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}tetrahydro-2H-pyran-2-carboxylic acid (non-preferred name), 1,2-ETHANEDIOL, DegT/DnrJ/EryC1/StrS aminotransferase, ... | | Authors: | Hofmeister, D.L, Seltzner, C.A, Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-23 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
8E75
 
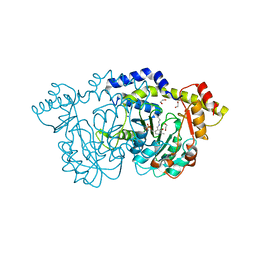 | | Crystal structure of Pcryo_0616, the aminotransferase required to synthesize UDP-N-acetyl-3-amino-D-glucosaminuronic acid (UDP-GlcNAc3NA) | | Descriptor: | 1,2-ETHANEDIOL, DegT/DnrJ/EryC1/StrS aminotransferase, SODIUM ION | | Authors: | Hofmeister, D.L, Seltzner, C.A, Bockhaus, N.J, thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-23 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
8E62
 
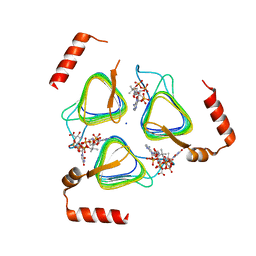 | | STRUCTURE OF Pcryo_0615 from Psychrobacter cryohalolentis, an N-acetyltransferase required to produce Diacetamido-2,3-dideoxy-D-glucuronic acid | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-4-amino-6-{[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxytetrahydro-2H-pyran-2-carboxylic acid, COENZYME A, SODIUM ION, ... | | Authors: | Hofmeister, D.L, Bockhaus, N.J, Seltzner, C.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-22 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
6B5E
 
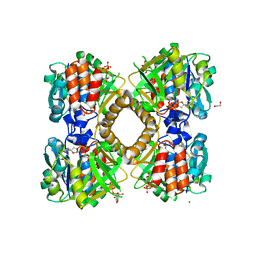 | | Mycobacterium tuberculosis RmlA in complex with dTDP-glucose | | Descriptor: | 1,2-ETHANEDIOL, 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, CHLORIDE ION, ... | | Authors: | Brown, H.A, Holden, H.M. | | Deposit date: | 2017-09-29 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of glucose-1-phosphate thymidylyltransferase from Mycobacterium tuberculosis reveals the location of an essential magnesium ion in the RmlA-type enzymes.
Protein Sci., 27, 2018
|
|
2R0T
 
 | |
6NBP
 
 | | Crystal Structure of a Sugar N-Formyltransferase from the Plant Pathogen Pantoea ananatis | | Descriptor: | N-formyltransferase, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, PHOSPHATE ION, ... | | Authors: | Hofmeister, D.L, Thoden, J.B, Holden, H.M. | | Deposit date: | 2018-12-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Investigation of a sugar N-formyltransferase from the plant pathogen Pantoea ananatis.
Protein Sci., 28, 2019
|
|
6OFU
 
 | | X-ray crystal structure of the YdjI aldolase from Escherichia coli K12 | | Descriptor: | CHLORIDE ION, YdjI aldolase, ZINC ION | | Authors: | Dopkins, B.J, Thoden, J.B, Huddleston, J.P, Narindoshvili, T, Fose, B, Rachel, F.M, Holden, H.M. | | Deposit date: | 2019-04-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Functional Characterization of YdjI, an Aldolase of Unknown Specificity inEscherichia coliK12.
Biochemistry, 58, 2019
|
|
3K5H
 
 | | Crystal structure of carboxyaminoimidazole ribonucleotide synthase from asperigillus clavatus complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Phosphoribosyl-aminoimidazole carboxylase | | Authors: | Thoden, J.B, Holden, H.M, Paritala, H, Firestine, S.M. | | Deposit date: | 2009-10-07 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of Aspergillus clavatus N(5)-carboxyaminoimidazole ribonucleotide synthetase
Biochemistry, 49, 2010
|
|
3K5I
 
 | | Crystal structure of N5-carboxyaminoimidazole synthase from aspergillus clavatus in complex with ADP and 5-aminoimadazole ribonucleotide | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Holden, H.M, Paritala, H, Firestine, S.M. | | Deposit date: | 2009-10-07 | | Release date: | 2009-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional studies of Aspergillus clavatus N(5)-carboxyaminoimidazole ribonucleotide synthetase
Biochemistry, 49, 2010
|
|
8DB5
 
 | | Crystal structure of the GDP-D-glycero-4-keto-d-lyxo-heptose-3,5-epimerase from Campylobacter jejuni, serotype HS:15 | | Descriptor: | CHLORIDE ION, GDP-D-glycero-4-keto-d-lyxo-heptose-3,5-epimerase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-14 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
8DCO
 
 | | Crystal structure of the GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase from Campylobacter jejuni, serotype HS:42 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase, ... | | Authors: | Thoden, J.B, Xiang, D.F, Ghosh, M.K, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
8DCL
 
 | | Crystal structure of the GDP-D-glycero-4-keto-D-lyxo-heptose-3-epimerase from campylobacter jejuni, serotype HS:23/36 | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-glycero-4-keto-D-lyxo-heptose-3-epimerase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-16 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
