6R8G
 
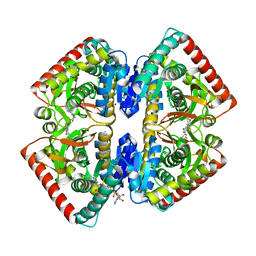 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with 4-(3,4-difluorophenyl)thiazol-2-amine | | Descriptor: | 4-[3,4-bis(fluoranyl)phenyl]-1,3-thiazol-2-amine, Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Fragment-Based Approach Identifies an Allosteric Pocket that Impacts Malate Dehydrogenase Activity
Commun Biol, 2021
|
|
6T2E
 
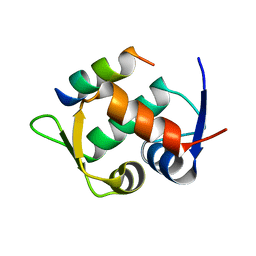 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, Stapled peptide GAR300-Gm | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6T2F
 
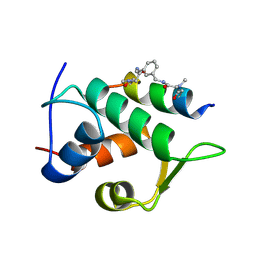 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | 2-(methylamino)-~{N}-[[3-[[2-(methylamino)ethanoylamino]methyl]phenyl]methyl]ethanamide, E3 ubiquitin-protein ligase Mdm2, MDM2 in complex with GAR300-Am | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6T2D
 
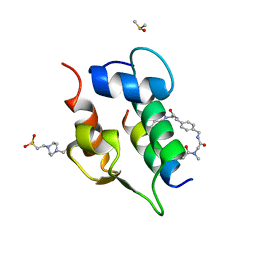 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | 2-(methylamino)-~{N}-[[4-[[2-(methylamino)ethanoylamino]methyl]phenyl]methyl]ethanamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1ACP
 
 | |
2D68
 
 | |
1B1U
 
 | |
2VID
 
 | | Serine protease SplB from Staphylococcus aureus at 1.8A resolution | | Descriptor: | SERINE PROTEASE SPLB | | Authors: | Dubin, G, Stec-Niemczyk, J, Kisielewska, M, Pustelny, K, Popowicz, G.M, Bista, M, Kantyka, T, Boulware, K.T, Stennicke, H.R, Czarna, A, Phopaisarn, M, Daugherty, P.S, Thogersen, I.B, Enghild, J.J, Thornberry, N, Dubin, A, Potempa, J. | | Deposit date: | 2007-11-30 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzymatic Activity of the Staphylococcus Aureus Splb Serine Protease is Induced by Substrates Containing the Sequence Trp-Glu-Leu-Gln.
J.Mol.Biol., 379, 2008
|
|
2W7U
 
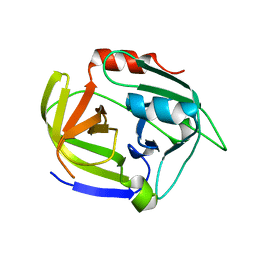 | | SplA serine protease of Staphylococcus aureus (2.4A) | | Descriptor: | SERINE PROTEASE SPLA | | Authors: | Stec-Niemczyka, J, Pustelny, K, Kisielewska, M, Bista, M, Boulware, K.T, Stennicke, H.R, Thogersen, I.B, Daugherty, P.S, Enghild, J.J, Popowicz, G.M, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2008-12-30 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and Functional Characterization of Spla, an Exclusively Specific Protease of Staphylococcus Aureus.
Biochem.J., 419, 2009
|
|
2W7S
 
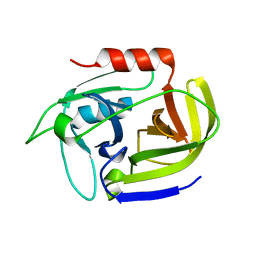 | | SplA serine protease of Staphylococcus aureus (1.8A) | | Descriptor: | SERINE PROTEASE SPLA | | Authors: | Stec-Niemczyka, J, Pustelny, K, Kisielewska, M, Bista, M, Boulware, K.T, Stennicke, H.R, Thogersen, I.B, Daugherty, P.S, Enghild, J.J, Popowicz, G.M, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2008-12-30 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Characterization of Spla, an Exclusively Specific Protease of Staphylococcus Aureus
Biochem.J., 419, 2009
|
|
2WSN
 
 | | Structure of Enhanced Cyan Fluorescent Protein at physiological pH | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Lelimousin, M, Noirclerc-Savoye, M, Lazareno-Saez, C, Paetzold, B, Le Vot, S, Chazal, R, Macheboeuf, P, Field, M.J, Bourgeois, D, Royant, A. | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Intrinsic Dynamics in Ecfp and Cerulean Control Fluorescence Quantum Yield.
Biochemistry, 48, 2009
|
|
1SVY
 
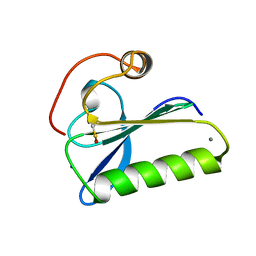 | | SEVERIN DOMAIN 2, 1.75 ANGSTROM CRYSTAL STRUCTURE | | Descriptor: | CALCIUM ION, SEVERIN, SODIUM ION | | Authors: | Puius, Y.A, Fedorov, E.V, Eichinger, L, Sullivan, M, Schleicher, M, Almo, S.C. | | Deposit date: | 1998-08-10 | | Release date: | 1999-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mapping the functional surface of domain 2 in the gelsolin superfamily.
Biochemistry, 39, 2000
|
|
1LDT
 
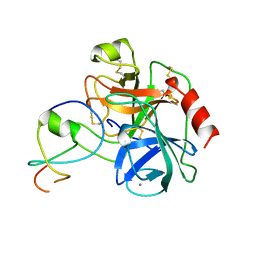 | | COMPLEX OF LEECH-DERIVED TRYPTASE INHIBITOR WITH PORCINE TRYPSIN | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPTASE INHIBITOR | | Authors: | Stubbs, M.T. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The three-dimensional structure of recombinant leech-derived tryptase inhibitor in complex with trypsin. Implications for the structure of human mast cell tryptase and its inhibition.
J.Biol.Chem., 272, 1997
|
|
1LU0
 
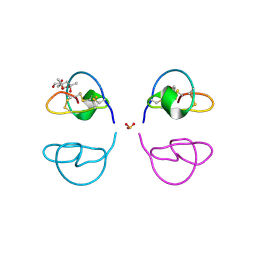 | | Atomic Resolution Structure of Squash Trypsin Inhibitor: Unexpected Metal Coordination | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Thaimattam, R, Tykarska, E, Bierzynski, A, Sheldrick, G.M, Jaskolski, M. | | Deposit date: | 2002-05-21 | | Release date: | 2002-08-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic resolution structure of squash trypsin inhibitor: unexpected metal coordination.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1PPE
 
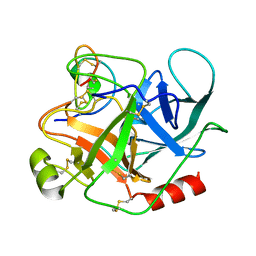 | |
