6R5J
 
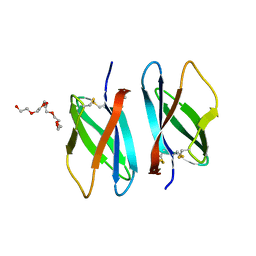 | |
1TUK
 
 | | Crystal structure of liganded type 2 non specific lipid transfer protein from wheat | | 分子名称: | 1-MYRISTOYL-2-HYDROXY-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL)], IODIDE ION, Nonspecific lipid-transfer protein 2G | | 著者 | Hoh, F, Pons, J.L, Gautier, M.F, De Lamotte, F, Dumas, C. | | 登録日 | 2004-06-25 | | 公開日 | 2005-04-05 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Structure of a liganded type 2 non-specific lipid-transfer protein from wheat and the molecular basis of lipid binding.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1JSG
 
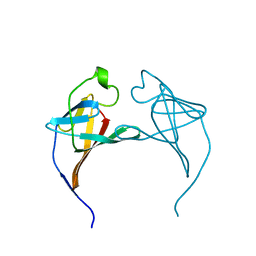 | | CRYSTAL STRUCTURE OF P14TCL1, AN ONCOGENE PRODUCT INVOLVED IN T-CELL PROLYMPHOCYTIC LEUKEMIA, REVEALS A NOVEL B-BARREL TOPOLOGY | | 分子名称: | ONCOGENE PRODUCT P14TCL1 | | 著者 | Hoh, F, Yang, Y.-S, Guignard, L, Padilla, A, Stern, R.-H, Lhoste, J.-M, Van Tilbeurgh, H. | | 登録日 | 1997-12-03 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of p14TCL1, an oncogene product involved in T-cell prolymphocytic leukemia, reveals a novel beta-barrel topology.
Structure, 6, 1998
|
|
3F6N
 
 | |
3F45
 
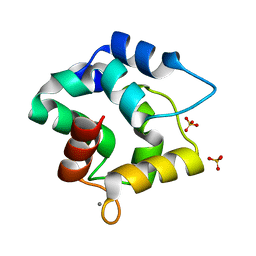 | |
1T7H
 
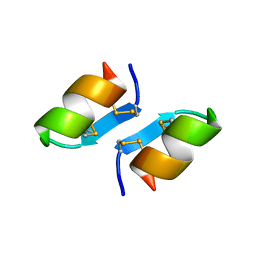 | | X-ray structure of [Lys(-2)-Arg(-1)-des(17-21)]-endothelin-1 peptide | | 分子名称: | Endothelin-1 | | 著者 | Hoh, F, Cerdan, R, Kaas, Q, Nishi, Y, Chiche, L, Kubo, S, Chino, N, Kobayashi, Y, Dumas, C, Aumelas, A. | | 登録日 | 2004-05-10 | | 公開日 | 2004-12-21 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | High-resolution X-ray structure of the unexpectedly stable dimer of the [Lys(-2)-Arg(-1)-des(17-21)]endothelin-1 peptide
Biochemistry, 43, 2004
|
|
7ZVM
 
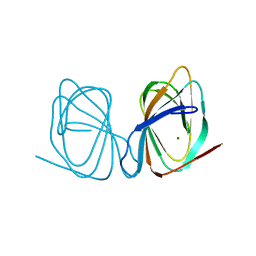 | |
7ZVY
 
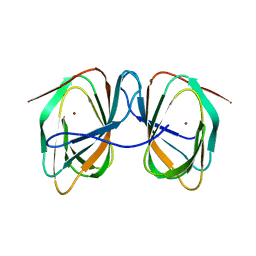 | |
8ACX
 
 | |
3K4T
 
 | |
2GFI
 
 | |
4L5M
 
 | |
4JMI
 
 | |
4JWL
 
 | |
4JMO
 
 | |
4JXH
 
 | |
6TY0
 
 | |
6TY2
 
 | |
4QFT
 
 | |
6XV2
 
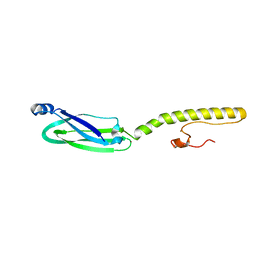 | | Full structure of RYMV P1 protein, derived from crystallographic and NMR data. | | 分子名称: | ZINC ION, p1 | | 著者 | Poignavent, V, Hoh, F, Vignols, F, Demene, H, Yang, Y, Gillet, F.X. | | 登録日 | 2020-01-21 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Flexible and Original Architecture of Two Unrelated Zinc Fingers Underlies the Role of the Multitask P1 in RYMV Spread.
J.Mol.Biol., 434, 2022
|
|
7P8U
 
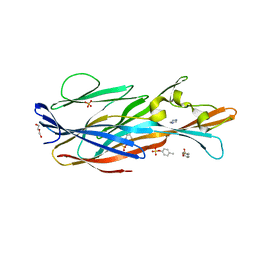 | | Crystal Structure of leukotoxin LukE from Staphylococcus aureus in complex with p-cresyl sulfate | | 分子名称: | (4-methylphenyl) hydrogen sulfate, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | 著者 | Lambey, P, Hoh, F, Peysson, F, Granier, S, Leyrat, C. | | 登録日 | 2021-07-23 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural insights into recognition of chemokine receptors by Staphylococcus aureus leukotoxins.
Elife, 11, 2022
|
|
7P8X
 
 | | Crystal Structure of leukotoxin LukE from Staphylococcus aureus in complex with a doubly sulfated CCR2 N-terminal peptide | | 分子名称: | C-C chemokine receptor type 2, IMIDAZOLE, Leucotoxin LukEv, ... | | 著者 | Lambey, P, Hoh, F, Peysson, F, Granier, S, Leyrat, C. | | 登録日 | 2021-07-23 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural insights into recognition of chemokine receptors by Staphylococcus aureus leukotoxins.
Elife, 11, 2022
|
|
7P93
 
 | | Crystal Structure of leukotoxin LukE from Staphylococcus aureus in complex with a sulfated ACKR1 N-terminal peptide | | 分子名称: | Atypical chemokine receptor 1, Leucotoxin LukEv | | 著者 | Lambey, P, Hoh, F, Peysson, F, Granier, S, Leyrat, C. | | 登録日 | 2021-07-23 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural insights into recognition of chemokine receptors by Staphylococcus aureus leukotoxins.
Elife, 11, 2022
|
|
7P8S
 
 | | Crystal Structure of leukotoxin LukE from Staphylococcus aureus at 1.9 Angstrom resolution | | 分子名称: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, HEXAETHYLENE GLYCOL, Leucotoxin LukEv, ... | | 著者 | Lambey, P, Hoh, F, Granier, S, Leyrat, C. | | 登録日 | 2021-07-23 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural insights into recognition of chemokine receptors by Staphylococcus aureus leukotoxins.
Elife, 11, 2022
|
|
7P8T
 
 | | Crystal Structure of leukotoxin LukE from Staphylococcus aureus at 1.5 Angstrom resolution | | 分子名称: | CHLORIDE ION, Leucotoxin LukEv | | 著者 | Lambey, P, Hoh, F, Granier, S, Leyrat, C. | | 登録日 | 2021-07-23 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.459 Å) | | 主引用文献 | Structural insights into recognition of chemokine receptors by Staphylococcus aureus leukotoxins.
Elife, 11, 2022
|
|
