7PIV
 
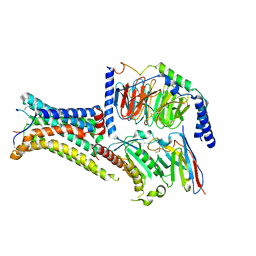 | | Active Melanocortin-4 receptor (MC4R)- Gs protein complex bound to agonist NDP-alpha-MSH at 2.86 A resolution. | | Descriptor: | CALCIUM ION, Camelid antibody VHH fragment - nanobody 35, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Heyder, N.A, Schmidt, A, Kleinau, G, Hilal, T, Scheerer, P. | | Deposit date: | 2021-08-23 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of active melanocortin-4 receptor-Gs-protein complexes with NDP-alpha-MSH and setmelanotide.
Cell Res., 31, 2021
|
|
4D67
 
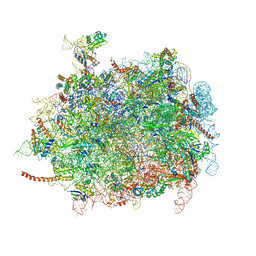 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | 28S RRNA, 5.8S RRNA, 5S RRNA, ... | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-04 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em Structures of Ribosomal 80S Complexes with Termination Factors and Cricket Paralysis Virus Ires Reveal the Ires in the Translocated State
Mol.Cell, 57, 2015
|
|
4D61
 
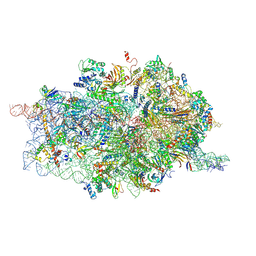 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-04 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
4D5N
 
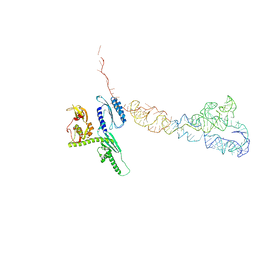 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | CRICKET PARALYSIS VIRUS IRES RNA, EUKARYOTIC PEPTIDE CHAIN RELEASE FACTOR SUBUNIT 1 | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-06 | | Release date: | 2015-02-04 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
4D5L
 
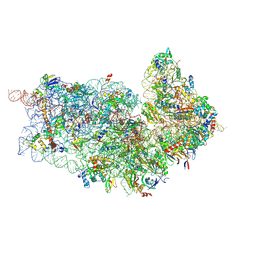 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | 18S RRNA 2, 40S RIBOSOMAL PROTEIN ES1, 40S RIBOSOMAL PROTEIN ES10, ... | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-02-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
4D5Y
 
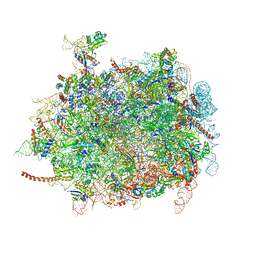 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | 28S Ribosomal RNA, 5.8S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em Structures of Ribosomal 80S Complexes with Termination Factors and Cricket Paralysis Virus Ires Reveal the Ires in the Translocated State
Mol.Cell, 57, 2015
|
|
8PTO
 
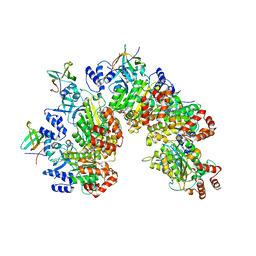 | | Structure of Rho pentamer in complex with Rof and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein rof, Transcription termination factor Rho | | Authors: | Said, N, Hilal, T, Wahl, M.C. | | Deposit date: | 2023-07-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Sm-like protein Rof inhibits transcription termination factor rho by binding site obstruction and conformational insulation.
Nat Commun, 15, 2024
|
|
8ALZ
 
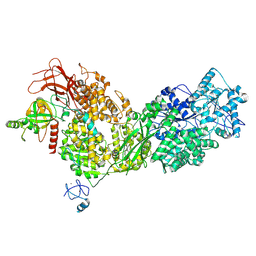 | | Cryo-EM structure of ASCC3 in complex with ASC1 | | Descriptor: | Activating signal cointegrator 1, Activating signal cointegrator 1 complex subunit 3, ZINC ION | | Authors: | Jia, J, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2022-08-01 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of ASCC3 in complex with ASC1
Nat Commun, 2023
|
|
6Z9T
 
 | | Transcription termination intermediate complex 5 | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6Z9Q
 
 | | Transcription termination intermediate complex 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6Z9P
 
 | | Transcription termination intermediate complex 1 | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6Z9S
 
 | | Transcription termination intermediate complex 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6Z9R
 
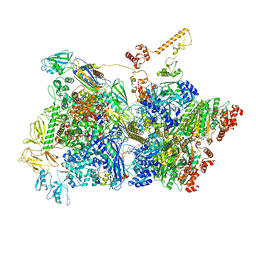 | | Transcription termination intermediate complex 3 | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7O6P
 
 | |
7O6Q
 
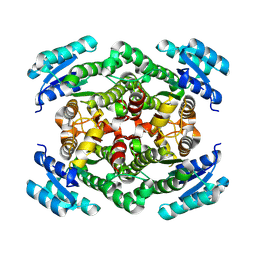 | |
7ADC
 
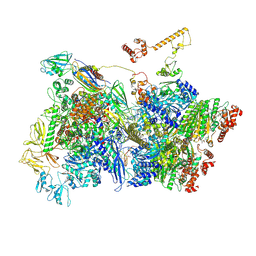 | | Transcription termination intermediate complex 3 delta NusG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-25 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7ADB
 
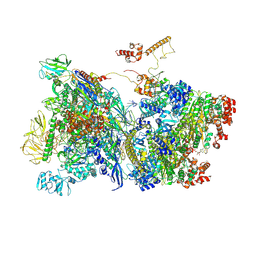 | | Transcription termination intermediate complex 1 delta NusG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7ADD
 
 | | Transcription termination intermediate complex IIIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-25 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7ADE
 
 | | Transcription termination complex IVa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-25 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6GBZ
 
 | | 50S ribosomal subunit assembly intermediate state 5 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
6GC8
 
 | | 50S ribosomal subunit assembly intermediate - 50S rec* | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-17 | | Release date: | 2018-06-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
6GC4
 
 | | 50S ribosomal subunit assembly intermediate state 3 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-17 | | Release date: | 2018-07-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
6GC0
 
 | | 50S ribosomal subunit assembly intermediate state 4 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
6GC6
 
 | | 50S ribosomal subunit assembly intermediate state 2 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-17 | | Release date: | 2018-07-04 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
6GC7
 
 | | 50S ribosomal subunit assembly intermediate state 1 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-17 | | Release date: | 2018-06-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
