4EWI
 
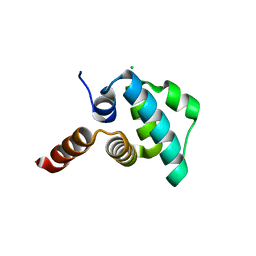 | | Crystal structure of the NLRP4 Pyrin domain | | Descriptor: | CHLORIDE ION, NACHT, LRR and PYD domains-containing protein 4, ... | | Authors: | Eibl, C, Hessenberger, M, Puehringer, S, Page, R, Diederichs, K, Peti, W. | | Deposit date: | 2012-04-27 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural and Functional Analysis of the NLRP4 Pyrin Domain.
Biochemistry, 51, 2012
|
|
4H1V
 
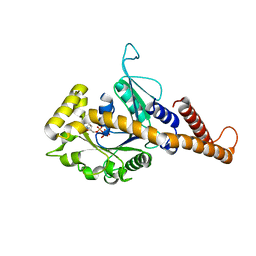 | | GMP-PNP bound dynamin-1-like protein GTPase-GED fusion | | Descriptor: | Dynamin-1-like protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Wenger, J, Klinglmayr, E, Eibl, C, Hessenberger, M, Goettig, P. | | Deposit date: | 2012-09-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Mapping of Human Dynamin-1-Like GTPase Domain Based on X-ray Structure Analyses.
Plos One, 8, 2013
|
|
6QL4
 
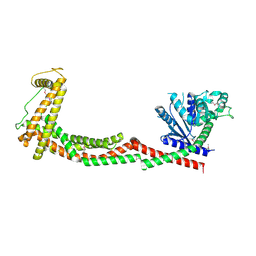 | | Crystal structure of nucleotide-free Mgm1 | | Descriptor: | 1,2-ETHANEDIOL, Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Wollweber, F, Pfitzner, A.-K, Muehleip, A, Sanchez, R, Kudryashev, M, Chiaruttin, N, Lilie, H, Schlegel, J, Rosenbaum, E, Hessenberger, M, Matthaeus, C, Noe, F, Roux, A, vanderLaan, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
4N1K
 
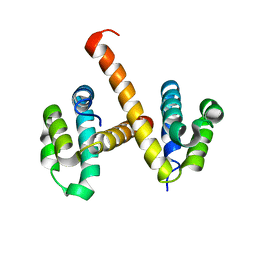 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N1J
 
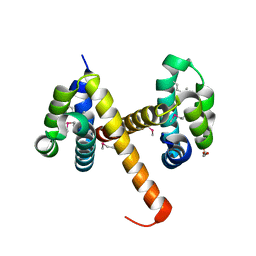 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | GLYCEROL, NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N1L
 
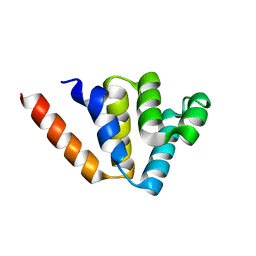 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7PV0
 
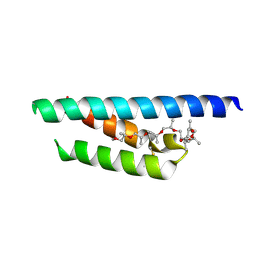 | | Crystal structure of a Mic60-Mic19 fusion protein | | Descriptor: | MICOS complex subunit MIC60,MICOS complex subunit MIC60-MIC19,Mic60-Mic19, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Funck, K, Bock-Bierbaum, T, Daumke, O. | | Deposit date: | 2021-10-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into crista junction formation by the Mic60-Mic19 complex.
Sci Adv, 8, 2022
|
|
7PV1
 
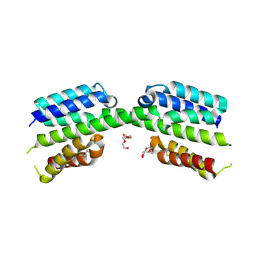 | |
7PUZ
 
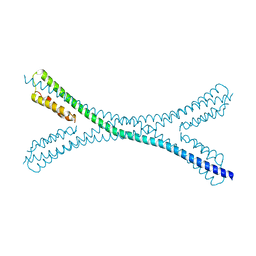 | |
4H1U
 
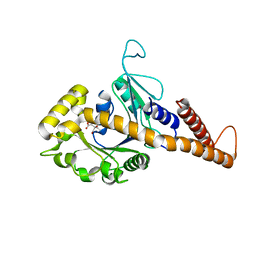 | | Nucleotide-free human dynamin-1-like protein GTPase-GED fusion | | Descriptor: | CITRATE ANION, Dynamin-1-like protein | | Authors: | Wenger, J, Klinglmayr, E, Puehringer, S, Goettig, P. | | Deposit date: | 2012-09-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Mapping of Human Dynamin-1-Like GTPase Domain Based on X-ray Structure Analyses.
Plos One, 8, 2013
|
|
6RZU
 
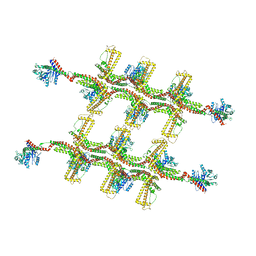 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZV
 
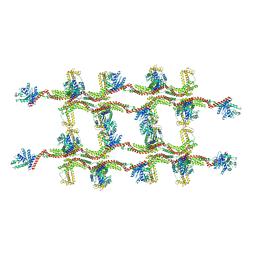 | | Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (20.6 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZW
 
 | | Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (18.799999 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZT
 
 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
4MP0
 
 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 2 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1003 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MOY
 
 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1953 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MOV
 
 | | 1.45 A Resolution Crystal Structure of Protein Phosphatase 1 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4503 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
