8GGZ
 
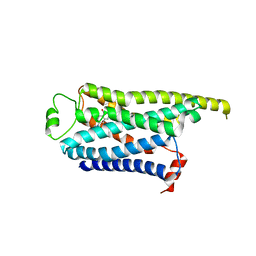 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #18 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 2024
|
|
7V2W
 
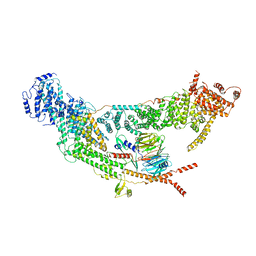 | | protomer structure from the dimer of yeast THO complex | | Descriptor: | Protein TEX1, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-10 | | Release date: | 2022-07-27 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
7V2Y
 
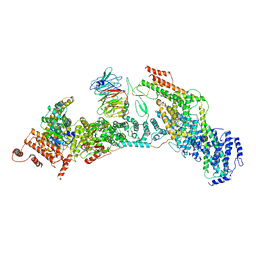 | | cryo-EM structure of yeast THO complex with Sub2 | | Descriptor: | ATP-dependent RNA helicase SUB2, Protein TEX1, THO complex subunit 2, ... | | Authors: | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-10 | | Release date: | 2022-07-27 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
7UL5
 
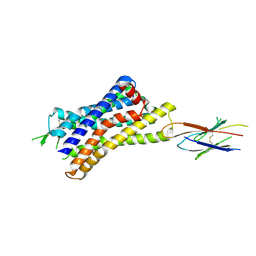 | |
7UL2
 
 | | CryoEM Structure of Inactive NTSR1 Bound to SR48692 and Nb6 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Nanobody 6, Neurotensin receptor 1, ... | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UL3
 
 | | CryoEM Structure of Inactive H2R Bound to Famotidine, Nb6M, and NabFab | | Descriptor: | Histamine H2 receptor, NabFab HC, NabFab LC, ... | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UL4
 
 | | CryoEM Structure of Inactive MOR Bound to Alvimopan and Mb6 | | Descriptor: | Megabody 6, Mu-type opioid receptor, N-[(2S)-2-{[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-yl]methyl}-3-phenylpropanoyl]glycine | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
