3WUE
 
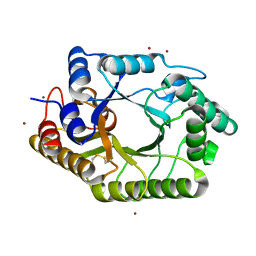 | | The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 | | 分子名称: | Endo-1,4-beta-xylanase A, ZINC ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | 著者 | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y, Guo, R.T. | | 登録日 | 2014-04-23 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
4WKR
 
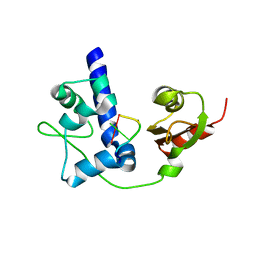 | | LaRP7 wrapping up the 3' hairpin of 7SK non-coding RNA (302-332) | | 分子名称: | 7SK GGHP4 (300-332), La-related protein 7 | | 著者 | Uchikawa, E, Natchiar, K.S, Han, X, Proux, F, Roblin, P, Zhang, E, Durand, A, Klaholz, B.P, Dock-Bregeon, A.-C. | | 登録日 | 2014-10-03 | | 公開日 | 2015-03-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7.
Nucleic Acids Res., 43, 2015
|
|
6H6H
 
 | |
6KY5
 
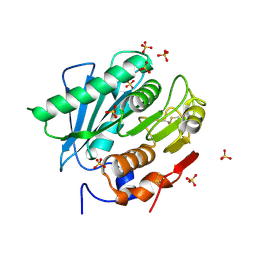 | | Crystal structure of a hydrolase mutant | | 分子名称: | PET hydrolase, SULFATE ION | | 著者 | Cui, Y.L, Chen, Y.C, Liu, X.Y, Dong, S.J, Han, J, Xiang, H, Chen, Q, Liu, H.Y, Han, X, Liu, W.D, Tang, S.Y, Wu, B. | | 登録日 | 2019-09-16 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.631 Å) | | 主引用文献 | Computational redesign of PETase for plasticbiodegradation by GRAPE strategy.
Biorxiv, 2020
|
|
8H8Y
 
 | | Crystal structure of AbHheG from Acidimicrobiia bacterium | | 分子名称: | GLYCEROL, alpha/beta hydrolase | | 著者 | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | 登録日 | 2022-10-24 | | 公開日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
8HQP
 
 | | Crystal structure of AbHheG mutant from Acidimicrobiia bacterium | | 分子名称: | AbHheG_m | | 著者 | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | 登録日 | 2022-12-13 | | 公開日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
7BWA
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 2 Insulin | | 分子名称: | Insulin fusion, Insulin receptor | | 著者 | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | 登録日 | 2020-04-14 | | 公開日 | 2021-04-14 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7CCR
 
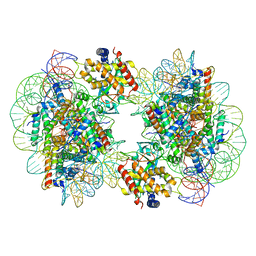 | | Structure of the 2:2 cGAS-nucleosome complex | | 分子名称: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | 著者 | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | 登録日 | 2020-06-17 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
7CCQ
 
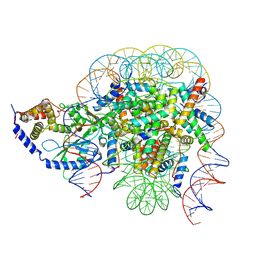 | | Structure of the 1:1 cGAS-nucleosome complex | | 分子名称: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | 著者 | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | 登録日 | 2020-06-17 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
8H83
 
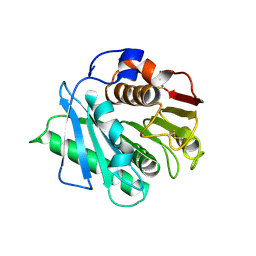 | | Crystal structure of a IsPETase variant V22 from Ideonella sakaiensis | | 分子名称: | Poly(ethylene terephthalate) hydrolase | | 著者 | Wei, H.L, Gao, S.F, Li, Q, Han, X, Gao, J, Liu, W.D. | | 登録日 | 2022-10-21 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Crystal structure of a IsPETase variant V22 from Ideonella sakaiensis
to be published
|
|
8IVP
 
 | | Crystal structure of MV in complex with LLP and FRU from Mycobacterium vanbaalenii | | 分子名称: | Branched chain amino acid: 2-keto-4-methylthiobutyrate aminotransferase, D-fructose | | 著者 | Li, Q, Zhu, Y.M, Gao, J, Wei, H.L, Han, X, Liu, W.D, Sun, Y.X. | | 登録日 | 2023-03-28 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Crystal structure of MV in complex with LLP and FRU from Mycobacterium vanbaalenii
To Be Published
|
|
8II9
 
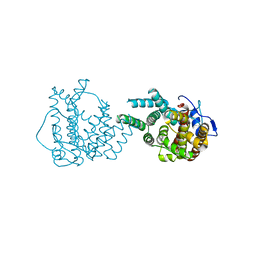 | | crystal structure of Hyp mutant from Hypoxylon sp. E7406B | | 分子名称: | Terpene synthase | | 著者 | Gao, J, Liu, W.D, Li, Q, Han, X, Wei, H.L, Dai, Z.J, Su, L.Q. | | 登録日 | 2023-02-24 | | 公開日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | crystal structure of Hyp
to be published
|
|
8IJT
 
 | | crystal structure of Hyp N135A mutant from Hypoxylon sp. E7406B | | 分子名称: | Terpene synthase | | 著者 | Gao, J, Su, L.Q, Li, Q, Han, X, Wei, H.L, Dai, Z.J, Liu, W.D. | | 登録日 | 2023-02-28 | | 公開日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | crystal structure of Hyp N135A mutant from Hypoxylon sp. E7406B
to be published
|
|
5YWW
 
 | | Archael RuvB-like Holiday junction helicase | | 分子名称: | GLYCEROL, Nucleotide binding protein PINc | | 著者 | Zhai, B, Yuan, Z, Han, X, DuPrez, K, Shen, Y, Fan, L. | | 登録日 | 2017-11-30 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | The archaeal ATPase PINA interacts with the helicase Hjm via its carboxyl terminal KH domain remodeling and processing replication fork and Holliday junction.
Nucleic Acids Res., 46, 2018
|
|
7XL5
 
 | | Crystal structure of the H42T/A85G/I86A mutant of a nadp-dependent alcohol dehydrogenase | | 分子名称: | NADP-dependent isopropanol dehydrogenase | | 著者 | Jiang, Y.Y, Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | 登録日 | 2022-04-21 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.604 Å) | | 主引用文献 | Crystal structure of the H42T/A85G/I86A mutant of a nadp-dependent alcohol dehydrogenase
To Be Published
|
|
7CUV
 
 | |
7E31
 
 | | Crystal structure of a novel alpha/beta hydrolase mutant in apo form | | 分子名称: | TRIETHYLENE GLYCOL, alpha/beta hydrolase | | 著者 | Gao, J, Han, X, Zheng, Y.Y, Liu, W.D. | | 登録日 | 2021-02-07 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7E30
 
 | | Crystal structure of a novel alpha/beta hydrolase in apo form in complex with citrate | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRIC ACID, SULFATE ION, ... | | 著者 | Gao, J, Han, X, Zheng, Y.Y, Liu, W.D. | | 登録日 | 2021-02-07 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7E9F
 
 | | Cryo-EM structure of the 2:1 Orc1 BAH domain in complex with nucleosome | | 分子名称: | DNA (147-mer), Histone H2A.2, Histone H2B.2, ... | | 著者 | Jiang, H, Yu, C, Liu, C.P, Han, X, Yu, Z, Xu, R.M. | | 登録日 | 2021-03-04 | | 公開日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Nucleosome binding relinquishes the association of the BAH domain of Orc1 with Sir1
To Be Published
|
|
7E9C
 
 | | Cryo-EM structure of the 1:1 Orc1 BAH domain in complex with nucleosome | | 分子名称: | DNA (147-mer), Histone H2A.2, Histone H2B.2, ... | | 著者 | Jiang, H, Yu, C, Liu, C.P, Han, X, Yu, Z, Xu, R.M. | | 登録日 | 2021-03-04 | | 公開日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Nucleosome binding relinquishes the association of the BAH domain of Orc1 with Sir1
To Be Published
|
|
7VPA
 
 | | Crystal structure of Ple629 from marine microbial consortium | | 分子名称: | hydrolase Ple629 | | 著者 | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | 登録日 | 2021-10-15 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
7VMD
 
 | | Crystal structure of a hydrolases Ple628 from marine microbial consortium | | 分子名称: | CALCIUM ION, hydrolase Ple628 | | 著者 | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | 登録日 | 2021-10-08 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
7VME
 
 | | Crystal structure of a hydrolase in apo form 2 | | 分子名称: | CALCIUM ION, hydrolase | | 著者 | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | 登録日 | 2021-10-08 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Crystal structure of a hydrolase in apo form 2
to be published
|
|
7VPB
 
 | | Crystal structure of a novel hydrolase in apo form | | 分子名称: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, ACETATE ION, plastic degrading hydrolase Ple629 | | 著者 | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | 登録日 | 2021-10-15 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural insight and engineering of a plastic degrading hydrolase Ple629.
Biochem.Biophys.Res.Commun., 626, 2022
|
|
7W6C
 
 | | Crystal structure of a PSH1 in complex with ligand J1K | | 分子名称: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | 著者 | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
