7LJP
 
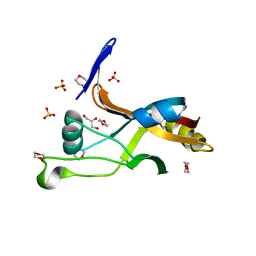 | | Structure of Thermotoga maritima SmpB | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2021-01-29 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Thermotoga maritima SmpB reveals its C-terminal tail domain in a helical conformation mimicking that of a ribosome-bound state
To Be Published
|
|
8I9M
 
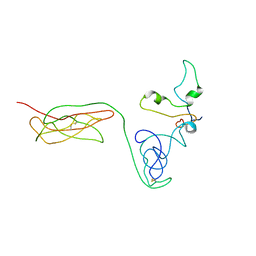 | | The RAGE and HMGB1 complex | | Descriptor: | Advanced glycosylation end product-specific receptor, High mobility group protein B1 | | Authors: | Han, C.W, Kim, H.J. | | Deposit date: | 2023-02-07 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (5.19 Å) | | Cite: | The RAGE and HMGB1 complex
To Be Published
|
|
8IZN
 
 | |
8HGJ
 
 | |
7C41
 
 | | KRAS G12V and H-REV107 peptide complex | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, HRAS-like suppressor 3, ... | | Authors: | Han, C.W, Jeong, M.S, Jang, S.B. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.276 Å) | | Cite: | A H-REV107 Peptide Inhibits Tumor Growth and Interacts Directly with Oncogenic KRAS Mutants.
Cancers (Basel), 12, 2020
|
|
7C3Z
 
 | |
7C40
 
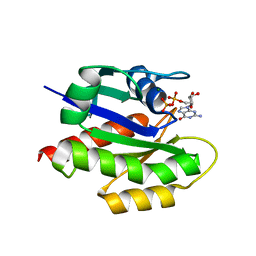 | | MgGDP bound KRAS G12V | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Han, C.W, Jeong, M.S, Jang, S.B. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.516 Å) | | Cite: | A H-REV107 Peptide Inhibits Tumor Growth and Interacts Directly with Oncogenic KRAS Mutants.
Cancers (Basel), 12, 2020
|
|
7YJJ
 
 | |
7DVD
 
 | | The crystal structure of p53 DNA binding domain and PUMA complex | | Descriptor: | Bcl-2-binding component 3, isoforms 1/2, Cellular tumor antigen p53, ... | | Authors: | Han, C.W, Lee, H.N, Jeong, M.S, Jang, S.B. | | Deposit date: | 2021-01-13 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis of the p53 DNA binding domain and PUMA complex.
Biochem.Biophys.Res.Commun., 548, 2021
|
|
7W5Q
 
 | |
6UGJ
 
 | |
6UGG
 
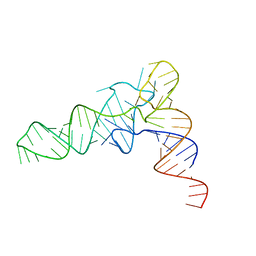 | | Structure of unmodified E. coli tRNA(Asp) | | Descriptor: | tRNAasp | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2019-09-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of an unmodified bacterial tRNA reveal intrinsic structural flexibility and plasticity as general properties of unbound tRNAs.
Rna, 26, 2020
|
|
6UGI
 
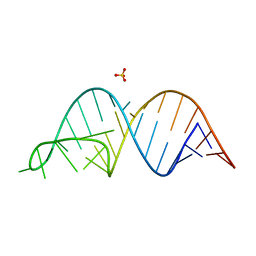 | |
6VMY
 
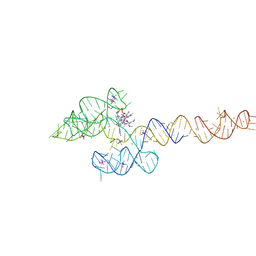 | | Structure of the B. subtilis cobalamin riboswitch | | Descriptor: | Adenosylcobalamin, B. subtilis cobalamin riboswitch, COBALT HEXAMMINE(III), ... | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of an atypical cobalamin riboswitch reveals RNA structural adaptability as basis for promiscuous ligand binding.
Nucleic Acids Res., 48, 2020
|
|
6CWX
 
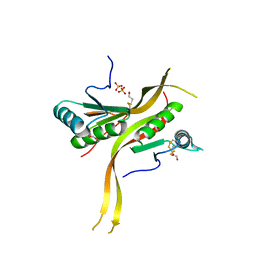 | | Crystal structure of human ribonuclease P/MRP proteins Rpp20/Rpp25 | | Descriptor: | FORMIC ACID, Ribonuclease P protein subunit p20, Ribonuclease P protein subunit p25, ... | | Authors: | Chan, C.W, Kiesel, B.R, Mondragon, A. | | Deposit date: | 2018-03-31 | | Release date: | 2018-04-18 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Human Rpp20/Rpp25 Reveals Quaternary Level Adaptation of the Alba Scaffold as Structural Basis for Single-stranded RNA Binding.
J. Mol. Biol., 430, 2018
|
|
8I9J
 
 | | The PKR and E3L complex | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase, RNA-binding protein E3 | | Authors: | Han, C.W, Kim, H.J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (6.39 Å) | | Cite: | Structural study of novel vaccinia virus E3L and dsRNA-dependent protein kinase complex.
Biochem.Biophys.Res.Commun., 665, 2023
|
|
5ZJD
 
 | | Lactate dehydrogenase with NADH and MLA | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase A chain, MALONATE ION | | Authors: | Han, C.W, Jang, S.B. | | Deposit date: | 2018-03-20 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Machilin A Inhibits Tumor Growth and Macrophage M2 Polarization Through the Reduction of Lactic Acid.
Cancers (Basel), 11, 2019
|
|
5ZJE
 
 | | LDHA-mla | | Descriptor: | L-lactate dehydrogenase A chain, MALONATE ION | | Authors: | Han, C.W, Jang, S.B. | | Deposit date: | 2018-03-20 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.929 Å) | | Cite: | Machilin A Inhibits Tumor Growth and Macrophage M2 Polarization Through the Reduction of Lactic Acid.
Cancers (Basel), 11, 2019
|
|
5ZJF
 
 | | LDHA-MA | | Descriptor: | 5,5'-[(2R,3S)-2,3-dimethylbutane-1,4-diyl]bis(2H-1,3-benzodioxole), L-lactate dehydrogenase A chain | | Authors: | Han, C.W, Jang, S.B. | | Deposit date: | 2018-03-20 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Machilin A Inhibits Tumor Growth and Macrophage M2 Polarization Through the Reduction of Lactic Acid.
Cancers (Basel), 11, 2019
|
|
4ZQA
 
 | |
5D90
 
 | |
5D8C
 
 | | Crystal structure of HiNmlR, a MerR family regulator lacking the sensor domain, bound to promoter DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*GP*AP*GP*TP*GP*AP*AP*CP*TP*CP*TP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*AP*GP*AP*GP*TP*TP*CP*AP*CP*TP*CP*TP*AP*AP*G)-3'), MerR family regulator protein | | Authors: | Counago, R.M, Chang, C.W, Chen, N.H, Djoko, K.Y, McEwan, A.G, Kobe, B. | | Deposit date: | 2015-08-17 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of thiol-based regulation of formaldehyde detoxification in H. influenzae by a MerR regulator with no sensor region.
Nucleic Acids Res., 44, 2016
|
|
7W5R
 
 | | KRAS G12V and peptide complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, LEU-TYR-ASP-VAL-ALA, ... | | Authors: | Kim, H.J, Han, C.W, Jang, S.B. | | Deposit date: | 2021-11-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.87 Å) | | Cite: | Structural basis of the oncogenic KRAS mutant and GJ101 complex.
Biochem.Biophys.Res.Commun., 641, 2023
|
|
5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | Descriptor: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | Authors: | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
5E01
 
 | | Crystal structure of HiNmlR, a MerR family regulator lacking the sensor domain, bound to palyndromic promoter DNA | | Descriptor: | 5'-D(*CP*TP*TP*AP*GP*AP*GP*TP*GP*CP*AP*CP*TP*CP*TP*AP*AP*G)-3', Uncharacterized HTH-type transcriptional regulator HI_0186 | | Authors: | Counago, R.M, Chang, C.W, Chen, N.H, Djoko, K.Y, McEwan, A.G, Kobe, B. | | Deposit date: | 2015-09-26 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of thiol-based regulation of formaldehyde detoxification in H. influenzae by a MerR regulator with no sensor region.
Nucleic Acids Res., 44, 2016
|
|
