8QVE
 
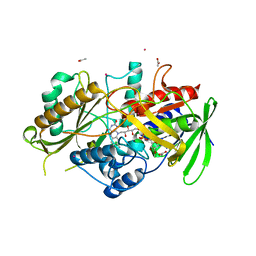 | | C-glucosyl oxidoreductase (DaCGO1) from Deinococcus aerius | | Descriptor: | ACETATE ION, CADMIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Furlanetto, V, Kalyani, D.C, Kostelac, A, Haltrich, D, Hallberg, B.M, Divne, C. | | Deposit date: | 2023-10-17 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characterization of a Gene Cluster Responsible for Deglycosylation of C-glucosyl Flavonoids and Xanthonoids by Deinococcus aerius.
J.Mol.Biol., 436, 2024
|
|
8QVC
 
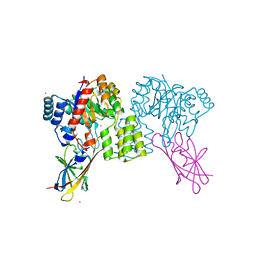 | | Deinococcus aerius TR0125 C-glucosyl deglycosidase (CGD), wild type crystal cryoprotected with glycerol | | Descriptor: | CADMIUM ION, DUF6379 domain-containing protein, Xylose isomerase-like TIM barrel domain-containing protein | | Authors: | Furlanetto, V, Kalyani, D.C, Kostelac, A, Haltrich, D, Hallberg, B.M, Divne, C. | | Deposit date: | 2023-10-17 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Characterization of a Gene Cluster Responsible for Deglycosylation of C-glucosyl Flavonoids and Xanthonoids by Deinococcus aerius.
J.Mol.Biol., 436, 2024
|
|
8QVD
 
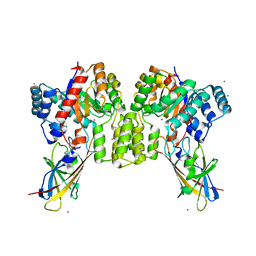 | | Deinococcus aerius TR0125 C-glucosyl deglycosidase (CGD), wild type crystal cryoprotected with glycerol | | Descriptor: | CADMIUM ION, DUF6379 domain-containing protein, Xylose isomerase-like TIM barrel domain-containing protein | | Authors: | Furlanetto, V, Kalyani, D.C, Kostelac, A, Haltrich, D, Hallberg, B.M, Divne, C. | | Deposit date: | 2023-10-17 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and Functional Characterization of a Gene Cluster Responsible for Deglycosylation of C-glucosyl Flavonoids and Xanthonoids by Deinococcus aerius.
J.Mol.Biol., 436, 2024
|
|
7NS6
 
 | |
8OJ6
 
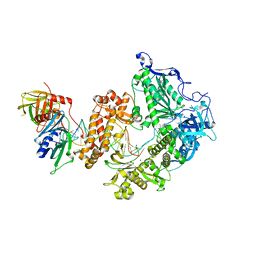 | | HSV-1 DNA polymerase-processivity factor complex in pre-translocation state | | Descriptor: | DNA (22-MER), DNA (48-MER), DNA polymerase catalytic subunit, ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
8OJ7
 
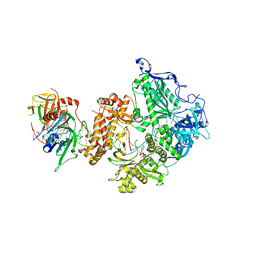 | | HSV-1 DNA polymerase-processivity factor complex in halted elongation state | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (53-MER), DNA (67-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
8OJC
 
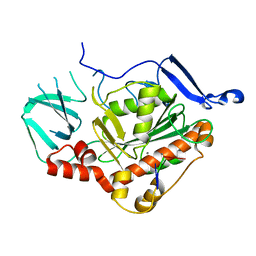 | | HSV-1 DNA polymerase active site in alternative exonuclease state | | Descriptor: | CALCIUM ION, DNA (47-MER), DNA polymerase catalytic subunit | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
8OJB
 
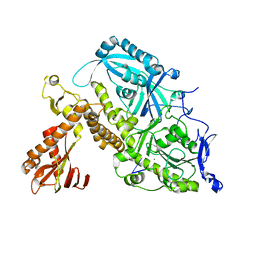 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state active site | | Descriptor: | CALCIUM ION, DNA (47-MER), DNA polymerase catalytic subunit | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
8P5M
 
 | |
8OJA
 
 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state | | Descriptor: | CALCIUM ION, DNA (47-MER), DNA (68-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (1.87 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
8OJD
 
 | | HSV-1 DNA polymerase beta-hairpin loop | | Descriptor: | CALCIUM ION, DNA (47-MER), DNA (68-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
4QI7
 
 | | Cellobiose dehydrogenase from Neurospora crassa, NcCDH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cellobiose dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
4QI4
 
 | | Dehydrogenase domain of Myriococcum thermophilum cellobiose dehydrogenase, MtDH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
4QI5
 
 | | Dehydrogenase domain of Myriococcum thermophilum cellobiose dehydrogenase with bound cellobionolactam, MtDH | | Descriptor: | (2R,3R,4R,5R)-4,5-dihydroxy-2-(hydroxymethyl)-6-oxopiperidin-3-yl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
4QI8
 
 | | Lytic polysaccharide monooxygenase 9F from Neurospora crassa, NcLPMO9F | | Descriptor: | COPPER (II) ION, Lytic polysaccharide monooxygenase, NITRATE ION | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
4QI3
 
 | | Cytochrome domain of Myriococcum thermophilum cellobiose dehydrogenase, MtCYT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cellobiose dehydrogenase, MAGNESIUM ION, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
4QI6
 
 | | Cellobiose dehydrogenase from Myriococcum thermophilum, MtCDH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cellobiose dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
3C5N
 
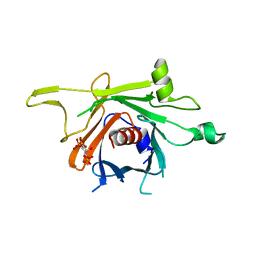 | | Structure of human TULP1 in complex with IP3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Tubby-related protein 1 | | Authors: | Busam, R.D, Lehtio, L, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van den Berg, S, Weigelt, J, Welin, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human TULP1 in complex with IP3
To be published
|
|
1QO7
 
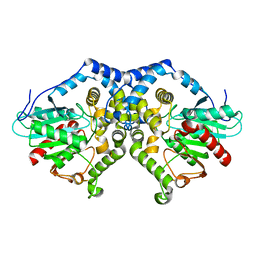 | | Structure of Aspergillus niger epoxide hydrolase | | Descriptor: | EPOXIDE HYDROLASE | | Authors: | Zou, J.-Y, Hallberg, B.M, Bergfors, T, Oesch, F, Arand, M, Mowbray, S.L, Jones, T.A. | | Deposit date: | 1999-11-04 | | Release date: | 2000-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Aspergillus Niger Epoxide Hydrolase at 1.8A Resolution: Implications for the Structure and Function of the Mammalian Microsomal Class of Epoxide Hydrolases
Structure, 8, 2000
|
|
5A2Y
 
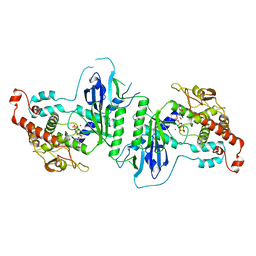 | | Crystal structure of mtPAP in complex with UTP | | Descriptor: | MAGNESIUM ION, MITOCHONDRIAL PROTEIN, URIDINE 5'-TRIPHOSPHATE | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A30
 
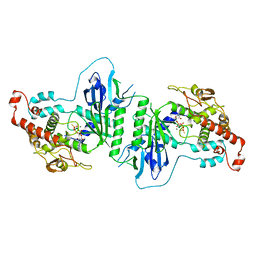 | | Crystal structure of mtPAP N472D mutant in complex with ATPgammaS | | Descriptor: | MAGNESIUM ION, MITOCHONDRIAL PROTEIN, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A2V
 
 | | Crystal structure of mtPAP in Apo form | | Descriptor: | CHLORIDE ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A2Z
 
 | | Crystal structure of mtPAP in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A2W
 
 | | Crystal structure of mtPAP in complex with ATPgammaS | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, MITOCHONDRIAL PROTEIN, ... | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A2X
 
 | | Crystal structure of mtPAP in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
