6TUT
 
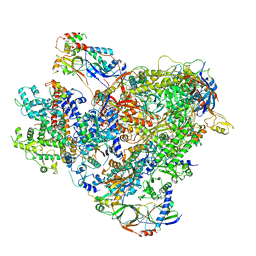 | | Cryo-EM structure of the RNA Polymerase III-Maf1 complex | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Vorlaender, M.K, Hagen, W.J.H, Mueller, C.W. | | Deposit date: | 2020-01-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis for RNA polymerase III transcription repression by Maf1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4UF8
 
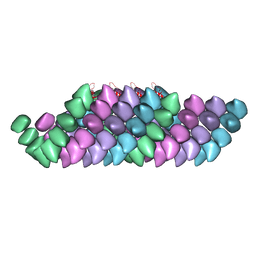 | | Electron cryo-microscopy structure of PB1-p62 filaments | | Descriptor: | SEQUESTOSOME-1 | | Authors: | Ciuffa, R, Lamark, T, Tarafder, A, Guesdon, A, Rybina, S, Hagen, W.J.H, Johansen, T, Sachse, C. | | Deposit date: | 2015-03-15 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | The Selective Autophagy Receptor P62 Forms a Flexible Filamentous Helical Scaffold.
Cell Rep., 11, 2015
|
|
4UDV
 
 | | Cryo-EM structure of TMV at 3.35 A resolution | | Descriptor: | 5'-D(*GP*AP*AP)-3', CAPSID PROTEIN | | Authors: | Fromm, S.A, Bharat, T.A.M, Jakobi, A.J, Hagen, W.J.H, Sachse, C. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Seeing Tobacco Mosaic Virus Through Direct Electron Detectors.
J.Struct.Biol., 189, 2015
|
|
4USN
 
 | | The structure of the immature HIV-1 capsid in intact virus particles at sub-nm resolution | | Descriptor: | P24 | | Authors: | Schur, F.K.M, Hagen, W.J.H, Rumlova, M, Ruml, T, Mueller, B, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structure of the Immature HIV-1 Capsid in Intact Virus Particles at 8.8 A Resolution.
Nature, 517, 2015
|
|
4UF9
 
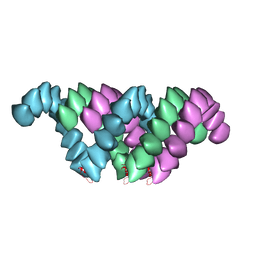 | | Electron cryo-microscopy structure of PB1-p62 type T filaments | | Descriptor: | SEQUESTOSOME-1 | | Authors: | Ciuffa, R, Lamark, T, Tarafder, A, Guesdon, A, Rybina, S, Hagen, W.J.H, Johansen, T, Sachse, C. | | Deposit date: | 2015-03-15 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | The Selective Autophagy Receptor P62 Forms a Flexible Filamentous Helical Scaffold.
Cell Rep., 11, 2015
|
|
6F42
 
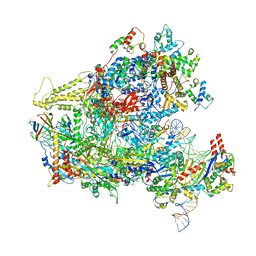 | | RNA Polymerase III closed complex CC1. | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Vorlaender, M.K, Khatter, H, Wetzel, R, Hagen, W.J.H, Mueller, C.W. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Molecular mechanism of promoter opening by RNA polymerase III.
Nature, 553, 2018
|
|
6F41
 
 | | RNA Polymerase III initially transcribing complex | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Vorlaender, M.K, Khatter, H, Wetzel, R, Hagen, W.J.H, Mueller, C.W. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular mechanism of promoter opening by RNA polymerase III.
Nature, 553, 2018
|
|
6F40
 
 | | RNA Polymerase III open complex | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Vorlaender, M.K, Khatter, H, Wetzel, R, Hagen, W.J.H, Mueller, C.W. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular mechanism of promoter opening by RNA polymerase III.
Nature, 553, 2018
|
|
6F44
 
 | | RNA Polymerase III closed complex CC2. | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Vorlaender, M.K, Khatter, H, Wetzel, R, Hagen, W.J.H, Mueller, C.W. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular mechanism of promoter opening by RNA polymerase III.
Nature, 553, 2018
|
|
7R5K
 
 | | Human nuclear pore complex (constricted) | | Descriptor: | Aladin, E3 SUMO-protein ligase RanBP2, Nuclear pore complex protein Nup107, ... | | Authors: | Mosalaganti, S, Obarska-Kosinska, A, Siggel, M, Taniguchi, R, Turonova, B, Zimmerli, C.E, Buczak, K, Schmidt, F.H, Margiotta, E, Mackmull, M.T, Hagen, W.J.H, Hummer, G, Kosinski, J, Beck, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | AI-based structure prediction empowers integrative structural analysis of human nuclear pores.
Science, 376, 2022
|
|
7R5J
 
 | | Human nuclear pore complex (dilated) | | Descriptor: | Aladin, E3 SUMO-protein ligase RanBP2, Nuclear pore complex protein Nup107, ... | | Authors: | Mosalaganti, S, Obarska-Kosinska, A, Siggel, M, Taniguchi, R, Turonova, B, Zimmerli, C.E, Buczak, K, Schmidt, F.H, Margiotta, E, Mackmull, M.T, Hagen, W.J.H, Hummer, G, Kosinski, J, Beck, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (50 Å) | | Cite: | AI-based structure prediction empowers integrative structural analysis of human nuclear pores
Science, 376, 2022
|
|
4COP
 
 | | HIV-1 capsid C-terminal domain mutant (Y169S) | | Descriptor: | CAPSID PROTEIN P24 | | Authors: | Bharat, T.A.M, Castillo-Menendez, L.R, Hagen, W.J.H, Lux, V, Igonet, S, Schorb, M, Schur, F.K.M, Krausslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2014-01-29 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cryo-Electron Microscopy of Tubular Arrays of HIV-1 Gag Resolves Structures Essential for Immature Virus Assembly.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4D1K
 
 | | Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. | | Descriptor: | GAG PROTEIN | | Authors: | Bharat, T.A.M, Castillo-Menendez, L.R, Hagen, W.J.H, Lux, V, Igonet, S, Schorb, M, Schur, F.K.M, Krauesslich, H.G, Briggs, J.A.G. | | Deposit date: | 2014-05-02 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Cryo-Electron Microscopy of Tubular Arrays of HIV-1 Gag Resolves Structures Essential for Immature Virus Assembly.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4COC
 
 | | HIV-1 capsid C-terminal domain mutant (Y169L) | | Descriptor: | CAPSID PROTEIN P24, SULFATE ION | | Authors: | Bharat, T.A.M, Castillo-Menendez, L.R, Hagen, W.J.H, Lux, V, Igonet, S, Schorb, M, Schur, F.K.M, Krausslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Cryo-Electron Microscopy of Tubular Arrays of HIV-1 Gag Resolves Structures Essential for Immature Virus Assembly.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5A9E
 
 | | Cryo-electron tomography and subtomogram averaging of Rous-Sarcoma- Virus deltaMBD virus-like particles | | Descriptor: | DELTAMBD GAG PROTEIN | | Authors: | Schur, F.K.M, Dick, R.A, Hagen, W.J.H, Vogt, V.M, Briggs, J.A.G. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | The Structure of Immature-Like Rous Sarcoma Virus Gag Particles Reveals a Structural Role for the P10 Domain in Assembly.
J.Virol., 89, 2015
|
|
5A1U
 
 | | The structure of the COPI coat triad | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
5A1Y
 
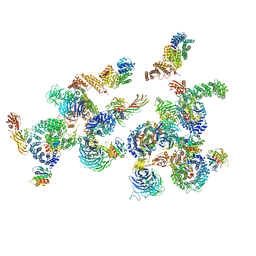 | | The structure of the COPI coat linkage IV | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
5AHV
 
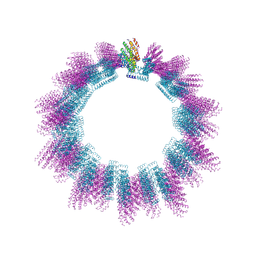 | | Cryo-EM structure of helical ANTH and ENTH tubules on PI(4,5)P2-containing membranes | | Descriptor: | ANTH DOMAIN OF ENDOCYTIC ADAPTOR SLA2, ENTH DOMAIN OF EPSIN ENT1 | | Authors: | Skruzny, M, Desfosses, A, Prinz, S, Dodonova, S.O, Gieras, A, Uetrecht, C, Jakobi, A.J, Abella, M, Hagen, W.J.H, Schulz, J, Meijers, R, Rybin, V, Briggs, J.A.G, Sachse, C, Kaksonen, M. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | An Organized Co-Assembly of Clathrin Adaptors is Essential for Endocytosis.
Dev.Cell, 33, 2015
|
|
5A1X
 
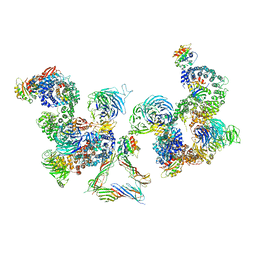 | | The structure of the COPI coat linkage III | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
5A1V
 
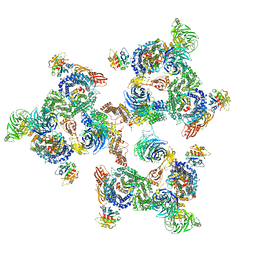 | | The structure of the COPI coat linkage I | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
5A1W
 
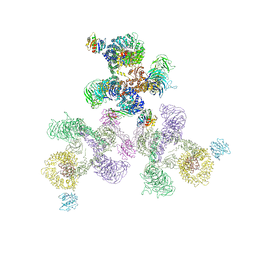 | | The structure of the COPI coat linkage II | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
5FJ8
 
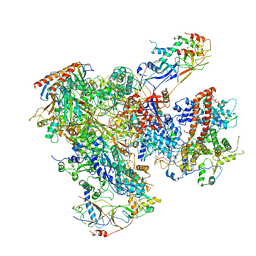 | | Cryo-EM structure of yeast RNA polymerase III elongation complex at 3. 9 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
5FJ9
 
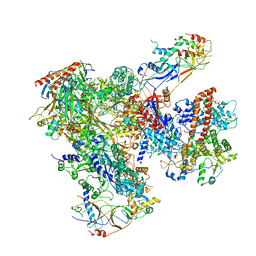 | | Cryo-EM structure of yeast apo RNA polymerase III at 4.6 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
5FJA
 
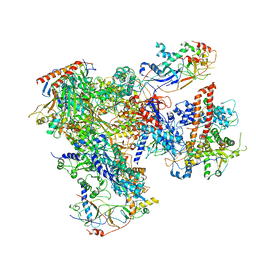 | | Cryo-EM structure of yeast RNA polymerase III at 4.7 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
5L93
 
 | | An atomic model of HIV-1 CA-SP1 reveals structures regulating assembly and maturation | | Descriptor: | Capsid protein p24 | | Authors: | Schur, F.K.M, Obr, M, Hagen, W.J.H, Wan, W, Arjen, J.J, Kirkpatrick, J.M, Sachse, C, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation.
Science, 353, 2016
|
|
