1DCK
 
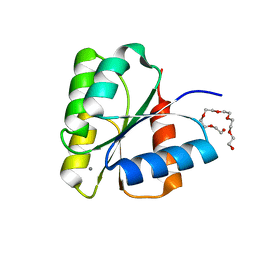 | | STRUCTURE OF UNPHOSPHORYLATED FIXJ-N COMPLEXED WITH MN2+ | | Descriptor: | MANGANESE (II) ION, POLYETHYLENE GLYCOL (N=34), TRANSCRIPTIONAL REGULATORY PROTEIN FIXJ | | Authors: | Gouet, P, Fabry, B, Guillet, V, Birck, C, Mourey, L, Kahn, D, Samama, J.P. | | Deposit date: | 1999-11-05 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural transitions in the FixJ receiver domain.
Structure Fold.Des., 7, 1999
|
|
1E25
 
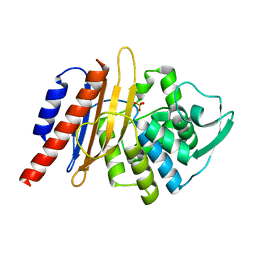 | | The high resolution structure of PER-1 class A beta-lactamase | | Descriptor: | EXTENDED-SPECTRUM BETA-LACTAMASE PER-1, SULFATE ION | | Authors: | Tranier, S, Bouthors, A.T, Maveyraud, L, Guillet, V, Sougakoff, W, Samama, J.P. | | Deposit date: | 2000-05-17 | | Release date: | 2000-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The High Resolution Crystal Structure for Class a Beta-Lactamase Per-1 Reveals the Bases for its Increase in Breadth of Activity
J.Biol.Chem., 275, 2000
|
|
1FFS
 
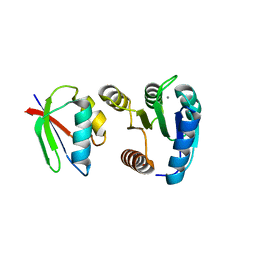 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY FROM CRYSTALS SOAKED IN ACETYL PHOSPHATE | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, MANGANESE (II) ION | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-26 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FFG
 
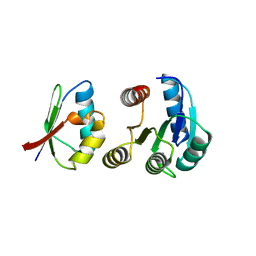 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY AT 2.1 A RESOLUTION | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, MANGANESE (II) ION | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-25 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FFW
 
 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY WITH A BOUND IMIDO DIPHOSPHATE | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, IMIDO DIPHOSPHATE, ... | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-26 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1DCM
 
 | | STRUCTURE OF UNPHOSPHORYLATED FIXJ-N WITH AN ATYPICAL CONFORMER (MONOMER A) | | Descriptor: | MAGNESIUM ION, TRANSCRIPTIONAL REGULATORY PROTEIN FIXJ | | Authors: | Gouet, P, Fabry, B, Guillet, V, Birck, C, Mourey, L, Kahn, D, Samama, J.P. | | Deposit date: | 1999-11-05 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural transitions in the FixJ receiver domain.
Structure Fold.Des., 7, 1999
|
|
5AJS
 
 | |
2JTG
 
 | | Solution structure of the THAP-zinc finger of THAP1 | | Descriptor: | THAP domain-containing protein 1, ZINC ION | | Authors: | Bessiere, D, Campagne, S, Milon, A, Gervais, V. | | Deposit date: | 2007-07-31 | | Release date: | 2007-12-11 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the THAP zinc finger of THAP1, a large C2CH DNA-binding module linked to Rb/E2F pathways
J.Biol.Chem., 283, 2008
|
|
7NBU
 
 | | Structure of the HigB1 toxin mutant K95A from Mycobacterium tuberculosis (Rv1955) and its target, the cspA mRNA, on the E. coli Ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Giudice, E, Mansour, M, Chat, S, D'Urso, G, Gillet, R, Genevaux, P. | | Deposit date: | 2021-01-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Substrate recognition and cryo-EM structure of the ribosome-bound TAC toxin of Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
5NJI
 
 | | Structure of the dehydratase domain of PpsC from Mycobacterium tuberculosis in complex with C12:1-CoA | | Descriptor: | Phthiocerol/phenolphthiocerol synthesis polyketide synthase type I PpsC, ~{S}-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (~{E})-dodec-2-enethioate | | Authors: | Gavalda, S, Faille, A, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2017-03-28 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into Substrate Modification by Dehydratases from Type I Polyketide Synthases.
J. Mol. Biol., 429, 2017
|
|
3CA4
 
 | | Sambucus nigra agglutinin II, tetragonal crystal form- complexed to lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
3CA5
 
 | | Crystal structure of Sambucus nigra agglutinin II (SNA-II)-tetragonal crystal form- complexed to alpha1 methylgalactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
3CAH
 
 | | Sambucus nigra aggutinin II. tetragonal crystal form- complexed to fucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-20 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
3CA1
 
 | | Sambucus nigra agglutinin II (SNA-II)- tetragonal crystal form- complexed to galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
3CA3
 
 | | Crystal structure of Sambucus Nigra Agglutinin II (SNA-II)-tetragonal crystal form- complexed to N-Acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
3CA6
 
 | | Sambucus nigra agglutinin II (SNA-II)- tetragonal crystal form- complexed to Tn antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
5I0K
 
 | |
4IYC
 
 | | Structure of the T244A mutant of the PANTON-VALENTINE LEUCOCIDIN component from STAPHYLOCOCCUS AUREUS | | Descriptor: | LukS-PV | | Authors: | Maveyraud, L, Guerin, F, Lavnetie, B.J, Prevost, G, Mourey, L. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Residues essential for panton-valentine leukocidin s component binding to its cell receptor suggest both plasticity and adaptability in its interaction surface
Plos One, 9, 2014
|
|
4J0O
 
 | |
4IYT
 
 | | Structure Of The Y184A Mutant Of The PANTON-VALENTINE LEUCOCIDIN S Component From STAPHYLOCOCCUS AUREUS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LukS-PV | | Authors: | Guerin, F, Laventie, B.J, Prevost, G, Mourey, L, Maveyraud, L. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Residues essential for panton-valentine leukocidin s component binding to its cell receptor suggest both plasticity and adaptability in its interaction surface
Plos One, 9, 2014
|
|
4IYA
 
 | | Structure of the Y250A mutant of the PANTON-VALENTINE LEUCOCIDIN S component from STAPHYLOCOCCUS AUREUS | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, LukS-PV | | Authors: | Maveyraud, L, Guerin, F, Laventie, B.J, Prevost, G, Mourey, L. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Residues essential for panton-valentine leukocidin s component binding to its cell receptor suggest both plasticity and adaptability in its interaction surface
Plos One, 9, 2014
|
|
4IZL
 
 | |
5L84
 
 | |
1B27
 
 | |
1B3S
 
 | |
