3U27
 
 | | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b | | 分子名称: | CALCIUM ION, GLYCEROL, Microcompartments protein, ... | | 著者 | Wu, R, Gu, M, Kerfeld, C.A, Salmeen, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-10-01 | | 公開日 | 2012-02-08 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.852 Å) | | 主引用文献 | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b
To be Published
|
|
3TSN
 
 | | 4-hydroxythreonine-4-phosphate dehydrogenase from Campylobacter jejuni | | 分子名称: | 4-hydroxythreonine-4-phosphate dehydrogenase, NICKEL (II) ION, UNKNOWN LIGAND | | 著者 | Osipiuk, J, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-13 | | 公開日 | 2011-10-12 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | 4-hydroxythreonine-4-phosphate dehydrogenase from Campylobacter jejuni.
To be Published
|
|
3UGS
 
 | | Crystal structure of a probable undecaprenyl diphosphate synthase (uppS) from Campylobacter jejuni | | 分子名称: | (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl dihydrogen phosphate, Undecaprenyl pyrophosphate synthase | | 著者 | Nocek, B, Gu, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-11-02 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.457 Å) | | 主引用文献 | Crystal structure of a probable undecaprenyl diphosphate synthase (uppS) from Campylobacter jejuni
TO BE PUBLISHED
|
|
7F03
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with ANP | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | 著者 | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | 登録日 | 2021-06-03 | | 公開日 | 2022-11-09 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F02
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | 著者 | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | 登録日 | 2021-06-03 | | 公開日 | 2022-11-09 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
4J1V
 
 | | Functional and structural studies of MOBKL1B, a Salvador/Warts/Hippo tumor suppressor pathway, in HCV replication | | 分子名称: | MOB kinase activator 1A, NS5A domain II peptide, ZINC ION | | 著者 | Chung, H.-Y, Gu, M, Rice, C.M. | | 登録日 | 2013-02-02 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Seed Sequence-Matched Controls Reveal Limitations of Small Interfering RNA Knockdown in Functional and Structural Studies of Hepatitis C Virus NS5A-MOBKL1B Interaction.
J.Virol., 88, 2014
|
|
4IYL
 
 | | 30S ribosomal protein S15 from Campylobacter jejuni | | 分子名称: | 30S ribosomal protein S15 | | 著者 | Osipiuk, J, Nocek, B, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-01-28 | | 公開日 | 2013-02-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | 30S ribosomal protein S15 from Campylobacter jejuni
To be Published
|
|
4IT5
 
 | | Chaperone HscB from Vibrio cholerae | | 分子名称: | CALCIUM ION, Co-chaperone protein HscB homolog | | 著者 | Osipiuk, J, Gu, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-01-17 | | 公開日 | 2013-01-30 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.152 Å) | | 主引用文献 | Chaperone HscB from Vibrio cholerae.
To be Published
|
|
4M0C
 
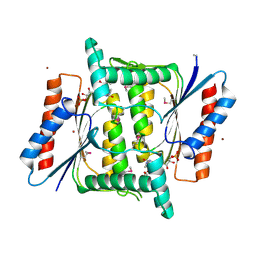 | | The crystal structure of a FMN-dependent NADH-azoreductase from Bacillus anthracis str. Ames Ancestor in complex with FMN. | | 分子名称: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, GLYCEROL, ... | | 著者 | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-08-01 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.073 Å) | | 主引用文献 | The crystal structure of a FMN-dependent NADH-azoreductase from Bacillus anthracis str. Ames Ancestor in complex with FMN.
To be Published
|
|
4NZP
 
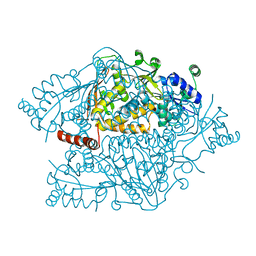 | | The crystal structure of argininosuccinate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | 分子名称: | Argininosuccinate synthase | | 著者 | Tan, K, Gu, M, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-12-12 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.307 Å) | | 主引用文献 | The crystal structure of argininosuccinate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
6B7J
 
 | |
3KYH
 
 | |
5YM3
 
 | | CYP76AH1-4pi from salvia miltiorrhiza | | 分子名称: | 4-PHENYL-1H-IMIDAZOLE, Ferruginol synthase, MANGANESE (II) ION, ... | | 著者 | Chang, Z. | | 登録日 | 2017-10-20 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | Crystal structure of CYP76AH1 in 4-PI-bound state from Salvia miltiorrhiza.
Biochem.Biophys.Res.Commun., 511, 2019
|
|
8I3U
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 14B1, Light chain of Fab 14B1, ... | | 著者 | Li, Z, Yu, F, Cao, S. | | 登録日 | 2023-01-18 | | 公開日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
8I3S
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain od Fab 7B3, Light chain of Fab 7B3, ... | | 著者 | Li, Z, Yu, F, Cao, S, ZHao, H. | | 登録日 | 2023-01-17 | | 公開日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
6VKL
 
 | |
1K6D
 
 | | CRYSTAL STRUCTURE OF ACETATE COA-TRANSFERASE ALPHA SUBUNIT | | 分子名称: | ACETATE COA-TRANSFERASE ALPHA SUBUNIT, MAGNESIUM ION | | 著者 | Korolev, S, Koroleva, O, Petterson, K, Collart, F, Dementieva, I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2001-10-15 | | 公開日 | 2002-06-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Autotracing of Escherichia coli acetate CoA-transferase alpha-subunit structure using 3.4 A MAD and 1.9 A native data.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8P8C
 
 | | HUMAN CD38 ECTODOMAIN BOUND TO COMPOUND 9-ADPR ADDUCT | | 分子名称: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[4-[[4-(2-methoxyethoxy)cyclohexyl]amino]-1-methyl-2-oxidanylidene-quinolin-6-yl]pyrazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Rangel, V, Zebisch, M, Doyle, K.J, Burli, R.W. | | 登録日 | 2023-05-31 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.653 Å) | | 主引用文献 | A Covalent Binding Mode of a Pyrazole-Based CD38 Inhibitor
Helv.Chim.Acta, 106, 2023
|
|
3GJZ
 
 | | Crystal structure of microcin immunity protein MccF from Bacillus anthracis str. Ames | | 分子名称: | Microcin immunity protein MccF | | 著者 | Nocek, B, Zhou, M, Kwon, K, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-03-09 | | 公開日 | 2009-04-14 | | 最終更新日 | 2012-07-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and Functional Characterization of Microcin C Resistance Peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
5CDM
 
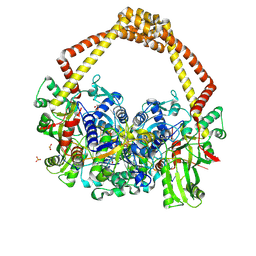 | | 2.5A structure of QPT-1 with S.aureus DNA gyrase and DNA | | 分子名称: | (2R,4S,4aS)-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4',6'(1'H,3'H)-trione, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | 著者 | Bax, B.D, Srikannathasan, V, Chan, P.F. | | 登録日 | 2015-07-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDN
 
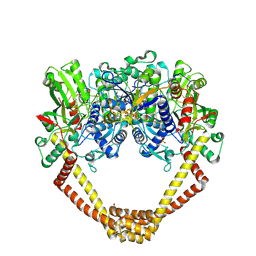 | | 2.8A structure of etoposide with S.aureus DNA gyrase and DNA | | 分子名称: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP**GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), ... | | 著者 | Bax, B.D, Srikannathasan, V, Chan, P.F. | | 登録日 | 2015-07-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDQ
 
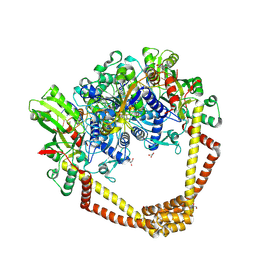 | | 2.95A structure of Moxifloxacin with S.aureus DNA gyrase and DNA | | 分子名称: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | 著者 | Bax, B.D, Srikannathasan, V, Chan, P.F. | | 登録日 | 2015-07-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2016-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDO
 
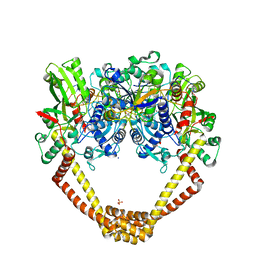 | | 3.15A structure of QPT-1 with S.aureus DNA gyrase and DNA | | 分子名称: | (2R,4S,4aS)-4',6'-dihydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidin]-2'-one, (2R,4S,4aS,5R)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, (2R,4S,4aS,5S)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, ... | | 著者 | Bax, B.D, Srikannathasan, V, Chan, P.F. | | 登録日 | 2015-07-04 | | 公開日 | 2015-12-16 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDR
 
 | | 2.65 structure of S.aureus DNA gyrase and artificially nicked DNA | | 分子名称: | DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*)-3'), DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | 著者 | Bax, B.D, Srikannathasan, V, Chan, P.F. | | 登録日 | 2015-07-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDP
 
 | | 2.45A structure of etoposide with S.aureus DNA gyrase and DNA | | 分子名称: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA gyrase subunit A, ... | | 著者 | Bax, B.D, Srikannathasan, V, Chan, P.F. | | 登録日 | 2015-07-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
