6YXE
 
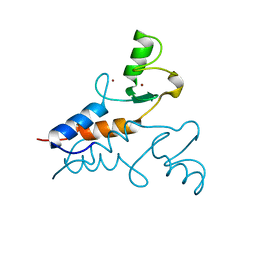 | | Structure of the Trim69 RING domain | | Descriptor: | E3 ubiquitin-protein ligase TRIM69, ZINC ION | | Authors: | Keown, J.R, Goldstone, D.C. | | Deposit date: | 2020-05-01 | | Release date: | 2020-10-14 | | Last modified: | 2020-12-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The RING domain of TRIM69 promotes higher-order assembly.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6BO1
 
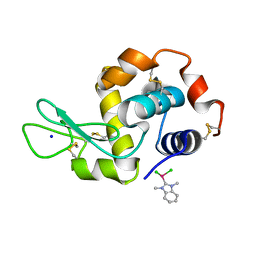 | | Mono-adduct formed after 3 days in the reaction of dichlorido(1,3-dimethylbenzimidazol-2-ylidene)(eta6-p-cymene)ruthenium(II) with HEWL | | Descriptor: | Lysozyme C, SODIUM ION, dichloro(1,3-dimethyl-1H-benzimidazol-3-ium-2-yl)ruthenium | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2017-11-17 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Unexpected arene ligand exchange results in the oxidation of an organoruthenium anticancer agent: the first X-ray structure of a protein-Ru(carbene) adduct.
Chem. Commun. (Camb.), 54, 2018
|
|
6BIQ
 
 | |
6BO2
 
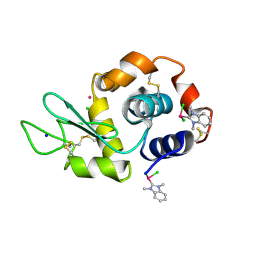 | | Adducts formed after 1 month in the reaction of dichlorido(1,3-dimethylbenzimidazol-2-ylidene)(eta6-p-cymene)ruthenium(II) with HEWL | | Descriptor: | CHLORIDE ION, Lysozyme C, RUTHENIUM ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2017-11-17 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Unexpected arene ligand exchange results in the oxidation of an organoruthenium anticancer agent: the first X-ray structure of a protein-Ru(carbene) adduct.
Chem. Commun. (Camb.), 54, 2018
|
|
6BIM
 
 | |
6BIO
 
 | |
6O5K
 
 | | Murine TRIM28 Bbox1 domain | | Descriptor: | Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Sun, Y, Keown, J.R, Goldstone, D.C. | | Deposit date: | 2019-03-03 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Dissection of Oligomerization by the TRIM28 Tripartite Motif and the Interaction with Members of the Krab-ZFP Family.
J.Mol.Biol., 431, 2019
|
|
6PL9
 
 | | Adduct formed after 1 month in the reaction of dichlorido(1,3-dimethylbenzimidaz ol-2-ylidene)(eta5-pentamethylcyclopentadienyl)rhodium(III) with HEWL | | Descriptor: | 2-(1-chloranyl-2,3,4,5,6-pentamethyl-1$l^{7}-rhodapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexan-1-yl)-1,3-dimethyl-benzimidazole, Lysozyme, SODIUM ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Probing the Paradigm of Promiscuity for N-Heterocyclic Carbene Complexes and their Protein Adduct Formation.
Angew.Chem.Int.Ed.Engl., 2021
|
|
6PLA
 
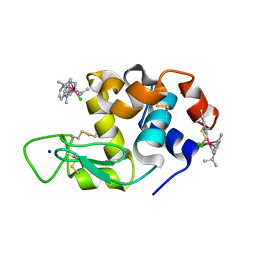 | | Adducts formed after 1 month in the reaction of dichlorido(1,3-dimethylbenzimidazol-2-ylidene)(eta6-p-cymene)osmium(II) with HEWL | | Descriptor: | Lysozyme, OSMIUM ION, SODIUM ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Probing the Paradigm of Promiscuity for N-Heterocyclic Carbene Complexes and their Protein Adduct Formation.
Angew.Chem.Int.Ed.Engl., 2021
|
|
6PLB
 
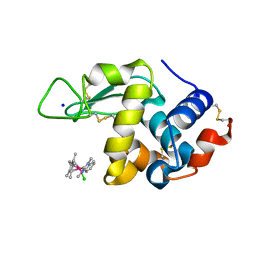 | | Adducts formed after 1 month in the reaction of dichlorido(1,3-dimethylbenzimida zol-2-ylidene)(eta5-pentamethylcyclopentadienyl)iridium(III) with HEWL | | Descriptor: | 2-(1-chloranyl-2,3,4,5,6-pentamethyl-1$l^{7}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexan-1-yl)-1,3-dimethyl-benzimidazole, Lysozyme, SODIUM ION | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing the Paradigm of Promiscuity for N-Heterocyclic Carbene Complexes and their Protein Adduct Formation.
Angew.Chem.Int.Ed.Engl., 2021
|
|
5UGJ
 
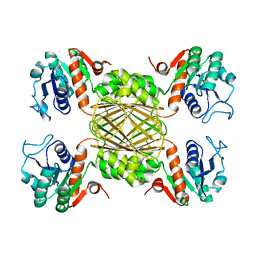 | |
5V4G
 
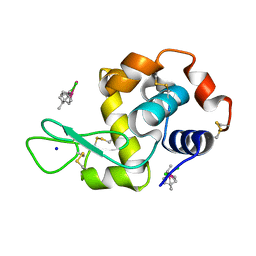 | | Ruthenium(II)(cymene)(chlorido)2-lysozyme adduct with two binding sites | | Descriptor: | Lysozyme C, PARA-CYMENE RUTHENIUM CHLORIDE, SODIUM ION | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The metalation of hen egg white lysozyme impacts protein stability as shown by ion mobility mass spectrometry, differential scanning calorimetry, and X-ray crystallography.
Chem. Commun. (Camb.), 53, 2017
|
|
5U5N
 
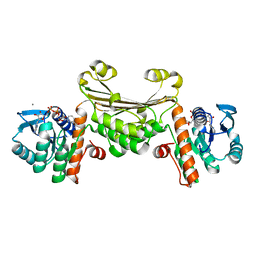 | |
5V4H
 
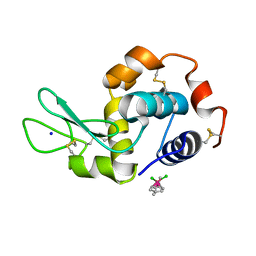 | | Ruthenium(II)(cymene)(chlorido)2-lysozyme adduct formed when ruthenium(II)(cymene)(bromido)2 underwent ligand exchange, resulting in one binding site | | Descriptor: | Lysozyme C, PARA-CYMENE RUTHENIUM CHLORIDE, SODIUM ION | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | The metalation of hen egg white lysozyme impacts protein stability as shown by ion mobility mass spectrometry, differential scanning calorimetry, and X-ray crystallography.
Chem. Commun. (Camb.), 53, 2017
|
|
5UA0
 
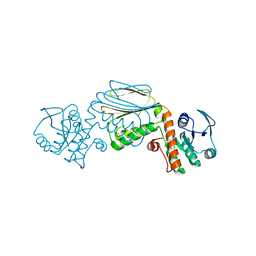 | |
5V4I
 
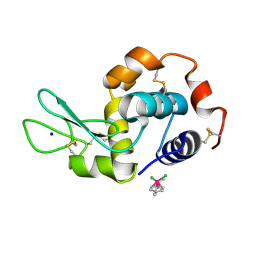 | | Osmium(II)(cymene)(chlorido)2-lysozyme adduct with one binding site | | Descriptor: | Lysozyme C, SODIUM ION, dichloro[(1,2,3,4,5,6-eta)-3-methyl-6-(propan-2-yl)benzene-1,2,4,5-tetrayl]osmium | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The metalation of hen egg white lysozyme impacts protein stability as shown by ion mobility mass spectrometry, differential scanning calorimetry, and X-ray crystallography.
Chem. Commun. (Camb.), 53, 2017
|
|
5U5I
 
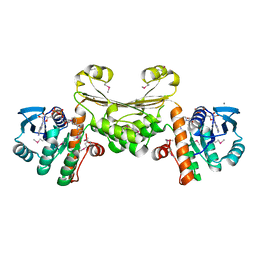 | |
2V4X
 
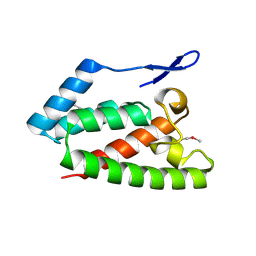 | | Crystal Structure of Jaagsiekte Sheep Retrovirus Capsid N-terminal domain | | Descriptor: | CAPSID PROTEIN P27 | | Authors: | Mortuza, G.B, Goldstone, D.C, Pashley, C, Haire, L.F, Palmarini, M, Taylor, W.R, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Capsid Amino-Terminal Domain from the Betaretrovirus, Jaagsiekte Sheep Retrovirus.
J.Mol.Biol., 386, 2009
|
|
2X1A
 
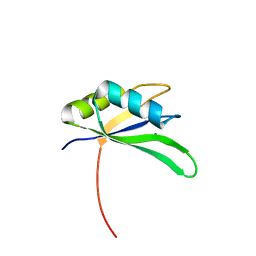 | | Structure of Rna15 RRM with RNA bound (G) | | Descriptor: | 5'-R(*GP*UP*UP*GP*UP)-3', MAGNESIUM ION, MRNA 3'-END-PROCESSING PROTEIN RNA15 | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2X1F
 
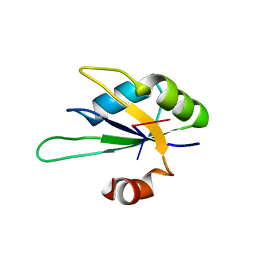 | | Structure of Rna15 RRM with bound RNA (GU) | | Descriptor: | 5'-R(*GP*UP*UP*GP*UP)-3', MRNA 3'-END-PROCESSING PROTEIN RNA15 | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2X1B
 
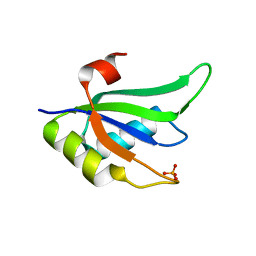 | | Structure of RNA15 RRM | | Descriptor: | MRNA 3'-END-PROCESSING PROTEIN RNA15, PHOSPHATE ION | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2XFV
 
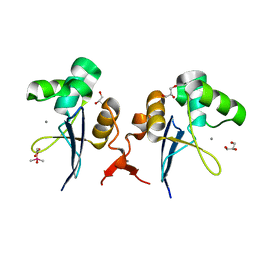 | | Structure of the amino-terminal domain from the cell-cycle regulator Swi6 | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Smerdon, S.J, Goldstone, D.C, Taylor, I.A. | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Amino-Terminal Domain from the Cell-Cycle Regulator Swi6S
Proteins, 78, 2010
|
|
3BP9
 
 | | Structure of B-tropic MLV capsid N-terminal domain | | Descriptor: | GLYCEROL, Gag protein, ISOPROPYL ALCOHOL | | Authors: | Gulnahar, M.B, Dodding, M.P, Goldstone, D.C, Haire, L.F, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2007-12-18 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of B-MLV capsid amino-terminal domain reveals key features of viral tropism, gag assembly and core formation
J.Mol.Biol., 376, 2008
|
|
6I28
 
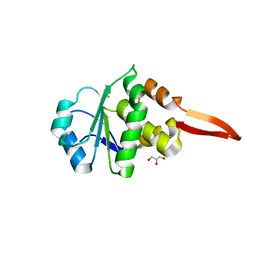 | | Crystal Structure of Cydia Pomonella PTP-2 phosphatase | | Descriptor: | CALCIUM ION, GLYCEROL, ORF98 PTP-2 | | Authors: | Huang, G, Keown, J.P, Oliver, M.R, Metcalf, P. | | Deposit date: | 2018-10-31 | | Release date: | 2019-02-20 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of protein tyrosine phosphatase-2 from Cydia pomonella granulovirus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
3U37
 
 | |
